Sodium »
PDB 5g3g-5gwa »
5gjo »
Sodium in PDB 5gjo: Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp
Enzymatic activity of Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp
All present enzymatic activity of Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp:
4.1.1.18;
4.1.1.18;
Protein crystallography data
The structure of Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp, PDB code: 5gjo
was solved by
H.-Y.Sagong,
K.-J.Kim,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 1.80 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 55.917, 122.859, 136.853, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.2 / 19.7 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp
(pdb code 5gjo). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 4 binding sites of Sodium where determined in the Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp, PDB code: 5gjo:
Jump to Sodium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Sodium where determined in the Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp, PDB code: 5gjo:
Jump to Sodium binding site number: 1; 2; 3; 4;
Sodium binding site 1 out of 4 in 5gjo
Go back to
Sodium binding site 1 out
of 4 in the Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp
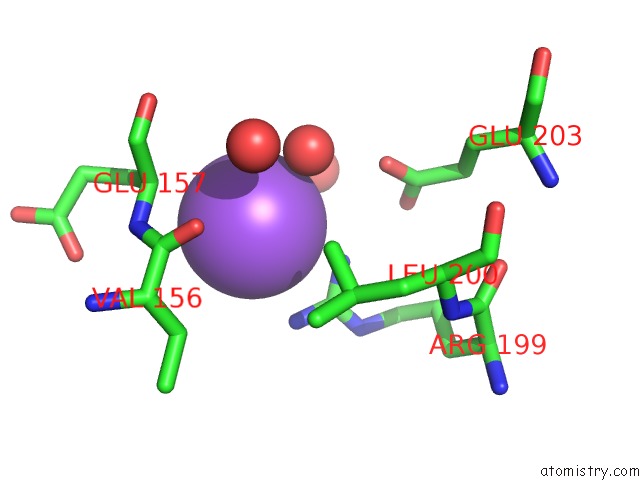
Mono view
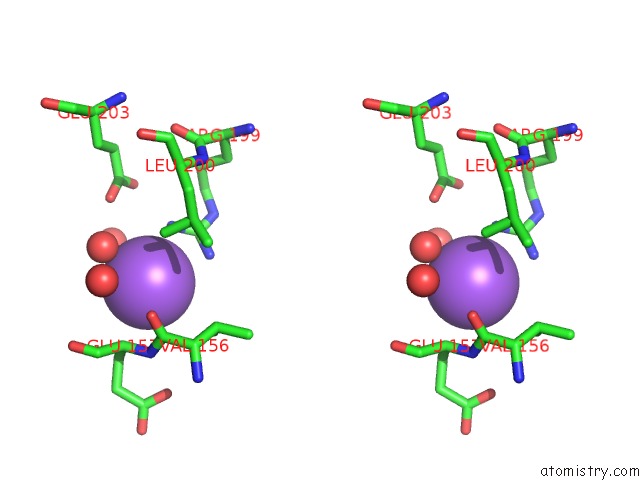
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp within 5.0Å range:
|
Sodium binding site 2 out of 4 in 5gjo
Go back to
Sodium binding site 2 out
of 4 in the Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp
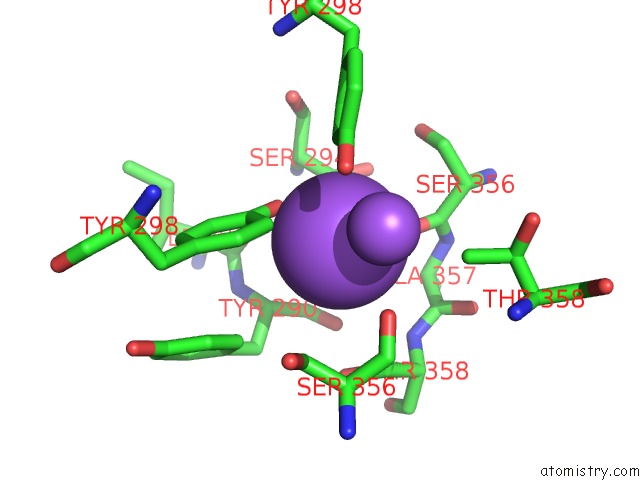
Mono view
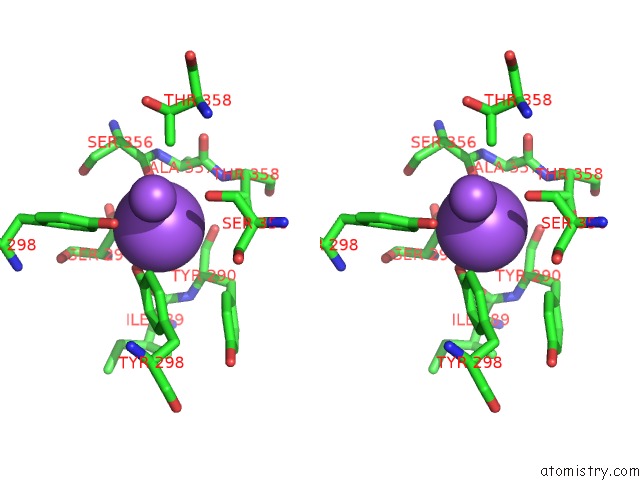
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp within 5.0Å range:
|
Sodium binding site 3 out of 4 in 5gjo
Go back to
Sodium binding site 3 out
of 4 in the Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp
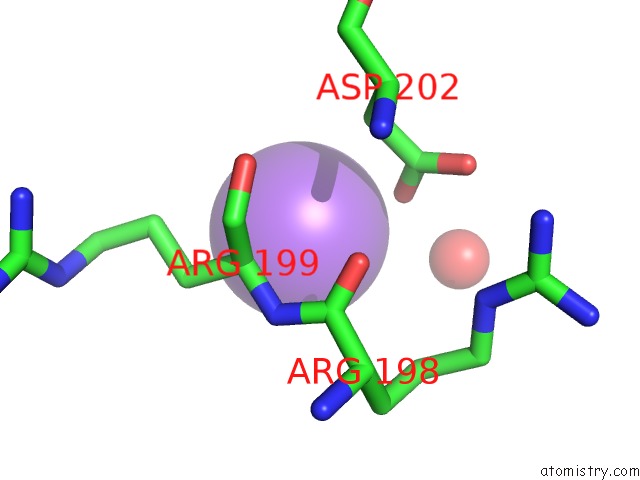
Mono view
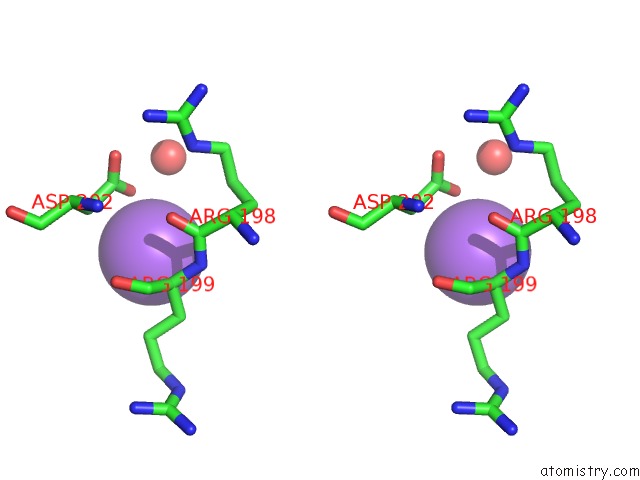
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp within 5.0Å range:
|
Sodium binding site 4 out of 4 in 5gjo
Go back to
Sodium binding site 4 out
of 4 in the Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp
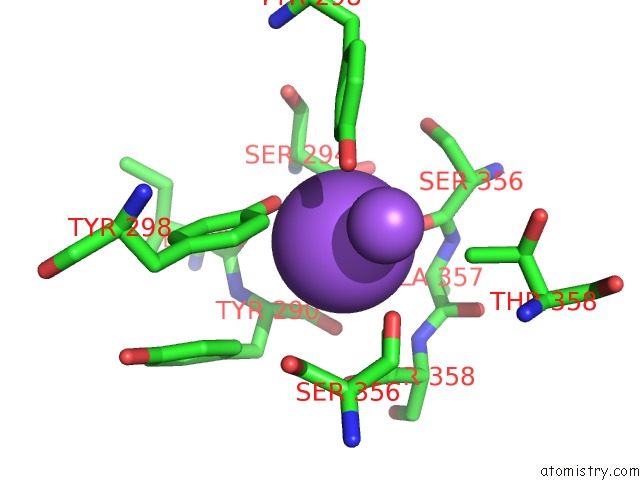
Mono view
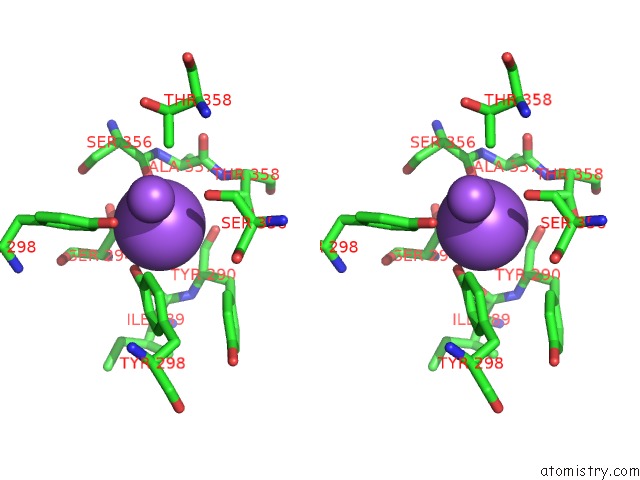
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Crystal Strcuture of Srldc Mutant (A225C/T302C) in Complex with Plp within 5.0Å range:
|
Reference:
H.-Y.Sagong,
K.-J.Kim.
Lysine Decarboxylase with An Enhanced Affinity For Pyridoxal 5-Phosphate By Disulfide Bond-Mediated Spatial Reconstitution Plos One V. 12 70163 2017.
ISSN: ESSN 1932-6203
PubMed: 28095457
DOI: 10.1371/JOURNAL.PONE.0170163
Page generated: Mon Oct 7 21:12:12 2024
ISSN: ESSN 1932-6203
PubMed: 28095457
DOI: 10.1371/JOURNAL.PONE.0170163
Last articles
Cl in 5FGECl in 5FGD
Cl in 5FGA
Cl in 5FG7
Cl in 5FF3
Cl in 5FF4
Cl in 5FDR
Cl in 5FF0
Cl in 5FF2
Cl in 5FEX