Sodium »
PDB 4wcn-4wxq »
4wfz »
Sodium in PDB 4wfz: Coxsackievirus B3 3DPOL Rna Dependent Rna Polymerase - Nacl Crystal Form
Enzymatic activity of Coxsackievirus B3 3DPOL Rna Dependent Rna Polymerase - Nacl Crystal Form
All present enzymatic activity of Coxsackievirus B3 3DPOL Rna Dependent Rna Polymerase - Nacl Crystal Form:
2.7.7.48;
2.7.7.48;
Protein crystallography data
The structure of Coxsackievirus B3 3DPOL Rna Dependent Rna Polymerase - Nacl Crystal Form, PDB code: 4wfz
was solved by
O.B.Peersen,
S.M.Mcdonald,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 58.88 / 1.80 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.423, 74.423, 288.757, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20 / 23.1 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Coxsackievirus B3 3DPOL Rna Dependent Rna Polymerase - Nacl Crystal Form
(pdb code 4wfz). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Coxsackievirus B3 3DPOL Rna Dependent Rna Polymerase - Nacl Crystal Form, PDB code: 4wfz:
In total only one binding site of Sodium was determined in the Coxsackievirus B3 3DPOL Rna Dependent Rna Polymerase - Nacl Crystal Form, PDB code: 4wfz:
Sodium binding site 1 out of 1 in 4wfz
Go back to
Sodium binding site 1 out
of 1 in the Coxsackievirus B3 3DPOL Rna Dependent Rna Polymerase - Nacl Crystal Form
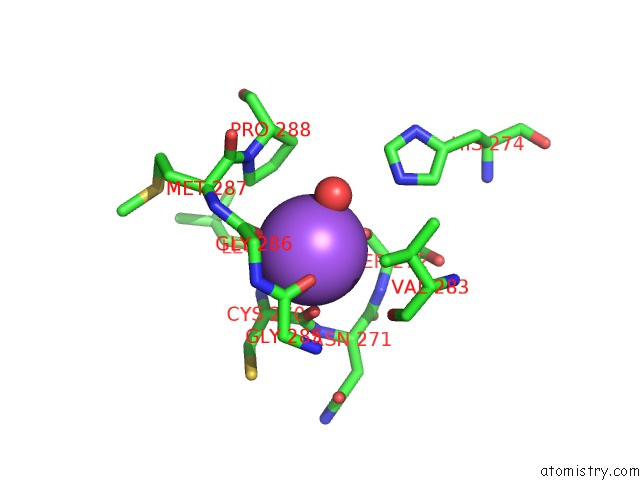
Mono view
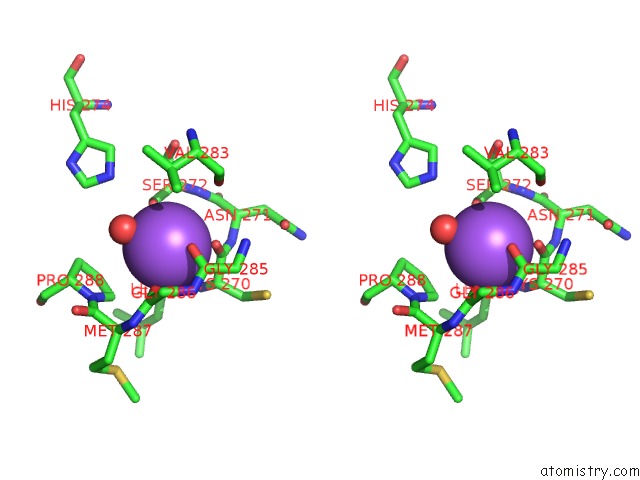
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Coxsackievirus B3 3DPOL Rna Dependent Rna Polymerase - Nacl Crystal Form within 5.0Å range:
|
Reference:
G.Campagnola,
S.Mcdonald,
S.Beaucourt,
M.Vignuzzi,
O.B.Peersen.
Structure-Function Relationships Underlying the Replication Fidelity of Viral Rna-Dependent Rna Polymerases. J.Virol. 2014.
ISSN: ESSN 1098-5514
PubMed: 25320316
DOI: 10.1128/JVI.01574-14
Page generated: Sun Aug 17 22:06:09 2025
ISSN: ESSN 1098-5514
PubMed: 25320316
DOI: 10.1128/JVI.01574-14
Last articles
Na in 6O5XNa in 6NZY
Na in 6O51
Na in 6O53
Na in 6O4Z
Na in 6O4Y
Na in 6NYQ
Na in 6O48
Na in 6O3C
Na in 6O3A