Sodium »
PDB 4mfc-4mzx »
4mxp »
Sodium in PDB 4mxp: Structural Basis For Pi(4)P-Specific Membrane Recruitment of the Legionella Pneumophila Effector Drra/Sidm
Protein crystallography data
The structure of Structural Basis For Pi(4)P-Specific Membrane Recruitment of the Legionella Pneumophila Effector Drra/Sidm, PDB code: 4mxp
was solved by
C.M.Del Campo,
A.K.Mishra,
Y.H.Wang,
C.R.Roy,
P.A.Janmey,
D.G.Lambright,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 26.90 / 1.83 |
| Space group | I 41 |
| Cell size a, b, c (Å), α, β, γ (°) | 128.552, 128.552, 53.222, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.9 / 20.8 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Structural Basis For Pi(4)P-Specific Membrane Recruitment of the Legionella Pneumophila Effector Drra/Sidm
(pdb code 4mxp). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Structural Basis For Pi(4)P-Specific Membrane Recruitment of the Legionella Pneumophila Effector Drra/Sidm, PDB code: 4mxp:
In total only one binding site of Sodium was determined in the Structural Basis For Pi(4)P-Specific Membrane Recruitment of the Legionella Pneumophila Effector Drra/Sidm, PDB code: 4mxp:
Sodium binding site 1 out of 1 in 4mxp
Go back to
Sodium binding site 1 out
of 1 in the Structural Basis For Pi(4)P-Specific Membrane Recruitment of the Legionella Pneumophila Effector Drra/Sidm

Mono view
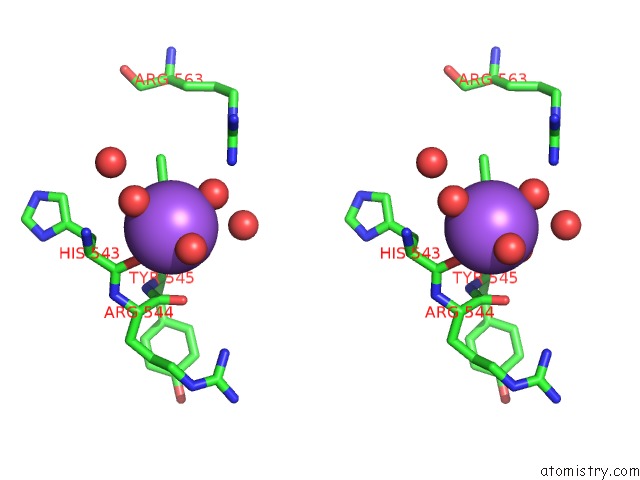
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Structural Basis For Pi(4)P-Specific Membrane Recruitment of the Legionella Pneumophila Effector Drra/Sidm within 5.0Å range:
|
Reference:
C.M.Del Campo,
A.K.Mishra,
Y.H.Wang,
C.R.Roy,
P.A.Janmey,
D.G.Lambright.
Structural Basis For Pi(4)P-Specific Membrane Recruitment of the Legionella Pneumophila Effector Drra/Sidm. Structure V. 22 397 2014.
ISSN: ISSN 0969-2126
PubMed: 24530282
DOI: 10.1016/J.STR.2013.12.018
Page generated: Sun Aug 17 20:42:00 2025
ISSN: ISSN 0969-2126
PubMed: 24530282
DOI: 10.1016/J.STR.2013.12.018
Last articles
Na in 6HWRNa in 6HY8
Na in 6HXI
Na in 6HUZ
Na in 6HTK
Na in 6HWP
Na in 6HUX
Na in 6HSX
Na in 6HRL
Na in 6HSN