Sodium »
PDB 4m04-4mfa »
4mao »
Sodium in PDB 4mao: RSK2 T493M C-Terminal Kinase Domain in Complex with RMM58
Enzymatic activity of RSK2 T493M C-Terminal Kinase Domain in Complex with RMM58
All present enzymatic activity of RSK2 T493M C-Terminal Kinase Domain in Complex with RMM58:
2.7.11.1;
2.7.11.1;
Protein crystallography data
The structure of RSK2 T493M C-Terminal Kinase Domain in Complex with RMM58, PDB code: 4mao
was solved by
R.M.Miller,
J.Taunton,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.12 / 2.60 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 47.500, 47.500, 288.500, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 23.5 / 29.2 |
Sodium Binding Sites:
The binding sites of Sodium atom in the RSK2 T493M C-Terminal Kinase Domain in Complex with RMM58
(pdb code 4mao). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the RSK2 T493M C-Terminal Kinase Domain in Complex with RMM58, PDB code: 4mao:
In total only one binding site of Sodium was determined in the RSK2 T493M C-Terminal Kinase Domain in Complex with RMM58, PDB code: 4mao:
Sodium binding site 1 out of 1 in 4mao
Go back to
Sodium binding site 1 out
of 1 in the RSK2 T493M C-Terminal Kinase Domain in Complex with RMM58
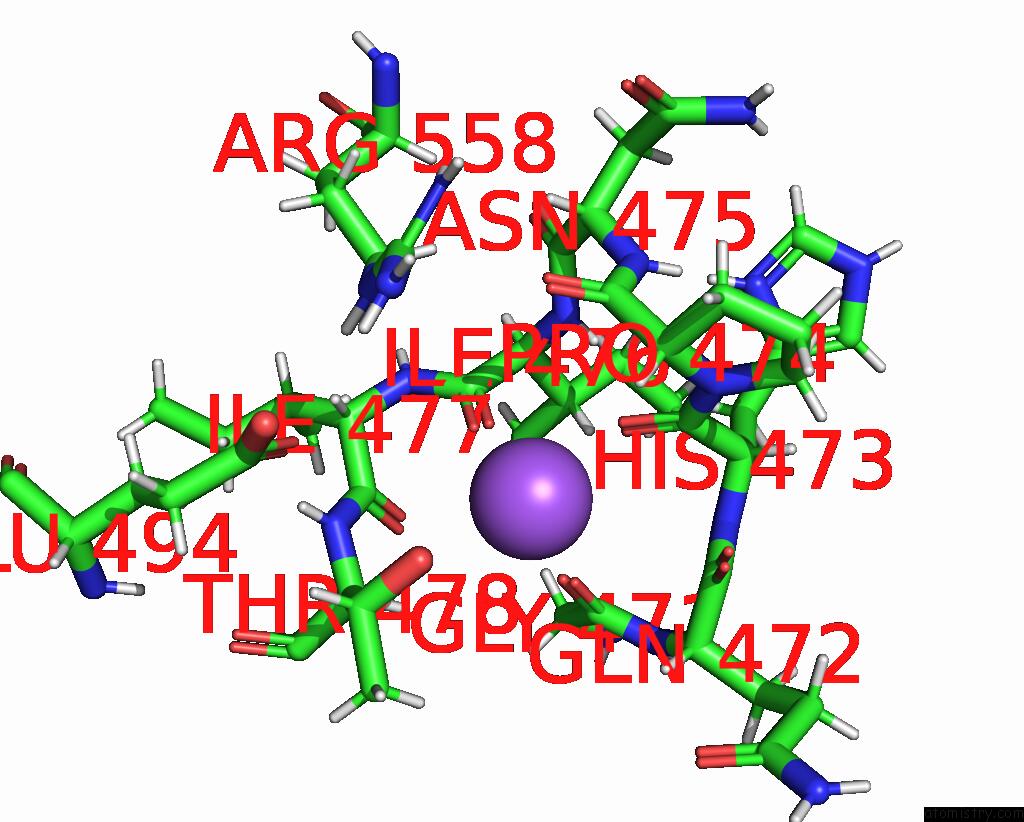
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of RSK2 T493M C-Terminal Kinase Domain in Complex with RMM58 within 5.0Å range:
|
Reference:
S.Krishnan,
R.M.Miller,
B.Tian,
R.D.Mullins,
M.P.Jacobson,
J.Taunton.
Design of Reversible, Cysteine-Targeted Michael Acceptors Guided By Kinetic and Computational Analysis. J.Am.Chem.Soc. V. 136 12624 2014.
ISSN: ISSN 0002-7863
PubMed: 25153195
DOI: 10.1021/JA505194W
Page generated: Mon Oct 7 16:58:01 2024
ISSN: ISSN 0002-7863
PubMed: 25153195
DOI: 10.1021/JA505194W
Last articles
Ca in 2XWGCa in 2XXL
Ca in 2XRC
Ca in 2XY8
Ca in 2XTS
Ca in 2XVT
Ca in 2XTA
Ca in 2XTT
Ca in 2XTJ
Ca in 2XT6