Sodium »
PDB 4jes-4jtk »
4jez »
Sodium in PDB 4jez: N79R Mutant of N-Acetylornithine Aminotransferase Complexed with L- Canaline
Enzymatic activity of N79R Mutant of N-Acetylornithine Aminotransferase Complexed with L- Canaline
All present enzymatic activity of N79R Mutant of N-Acetylornithine Aminotransferase Complexed with L- Canaline:
2.6.1.11; 2.6.1.17;
2.6.1.11; 2.6.1.17;
Protein crystallography data
The structure of N79R Mutant of N-Acetylornithine Aminotransferase Complexed with L- Canaline, PDB code: 4jez
was solved by
S.Bisht,
S.R.Bharath,
M.R.N.Murthy,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 28.01 / 1.55 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 96.600, 112.030, 65.110, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.7 / 17.6 |
Sodium Binding Sites:
The binding sites of Sodium atom in the N79R Mutant of N-Acetylornithine Aminotransferase Complexed with L- Canaline
(pdb code 4jez). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the N79R Mutant of N-Acetylornithine Aminotransferase Complexed with L- Canaline, PDB code: 4jez:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the N79R Mutant of N-Acetylornithine Aminotransferase Complexed with L- Canaline, PDB code: 4jez:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 4jez
Go back to
Sodium binding site 1 out
of 2 in the N79R Mutant of N-Acetylornithine Aminotransferase Complexed with L- Canaline
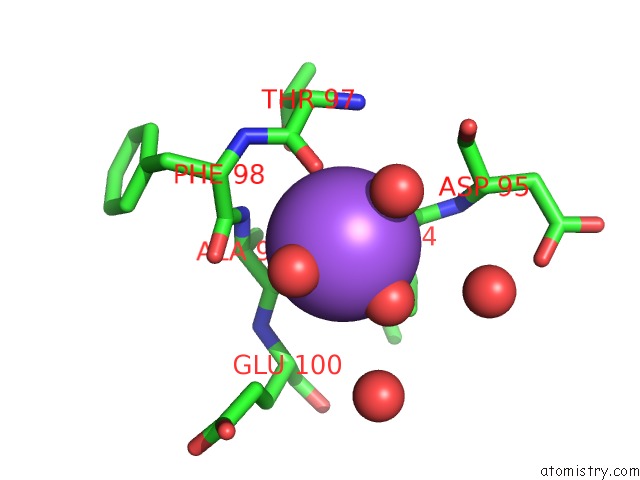
Mono view
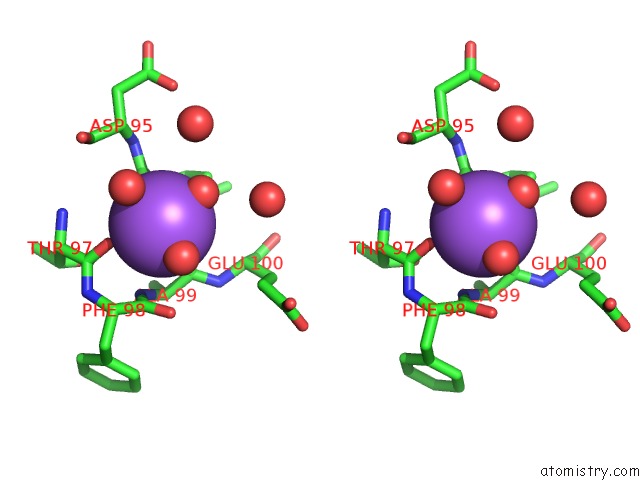
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of N79R Mutant of N-Acetylornithine Aminotransferase Complexed with L- Canaline within 5.0Å range:
|
Sodium binding site 2 out of 2 in 4jez
Go back to
Sodium binding site 2 out
of 2 in the N79R Mutant of N-Acetylornithine Aminotransferase Complexed with L- Canaline
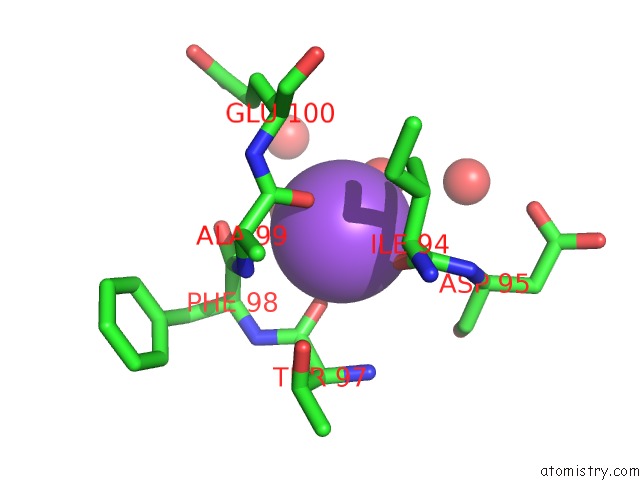
Mono view
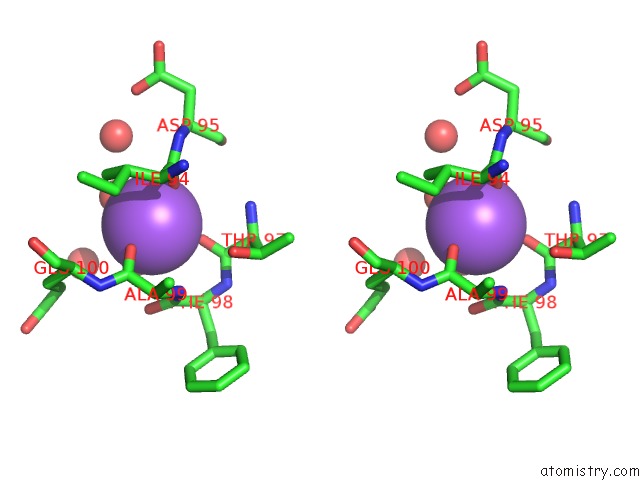
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of N79R Mutant of N-Acetylornithine Aminotransferase Complexed with L- Canaline within 5.0Å range:
|
Reference:
S.Bisht,
S.R.Bharath,
M.R.N.Murthy.
Conformational Transitions, Ligand Specificity and Catalysis in N-Acetylornithine Aminotransferase: Implications on Drug Designing and Rational Enzyme Engineering in Omega Aminotransferases To Be Published.
Page generated: Sun Aug 17 20:03:36 2025
Last articles
Na in 6TS4Na in 6TRR
Na in 6TQD
Na in 6TQQ
Na in 6TQP
Na in 6TQB
Na in 6TP2
Na in 6TP0
Na in 6TP1
Na in 6TQ4