Sodium »
PDB 4coo-4ddo »
4cte »
Sodium in PDB 4cte: Crystal Structure of the Catalytic Domain of the Modular Laminarinase Zglamc Mutant E142S in Complex with A Thio- Oligosaccharide
Enzymatic activity of Crystal Structure of the Catalytic Domain of the Modular Laminarinase Zglamc Mutant E142S in Complex with A Thio- Oligosaccharide
All present enzymatic activity of Crystal Structure of the Catalytic Domain of the Modular Laminarinase Zglamc Mutant E142S in Complex with A Thio- Oligosaccharide:
3.2.1.39;
3.2.1.39;
Protein crystallography data
The structure of Crystal Structure of the Catalytic Domain of the Modular Laminarinase Zglamc Mutant E142S in Complex with A Thio- Oligosaccharide, PDB code: 4cte
was solved by
A.Labourel,
M.Jam,
L.Legentil,
B.Sylla,
E.Ficko-Blean,
J.H.Hehemann,
V.Ferrieres,
M.Czjzek,
G.Michel,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.11 / 1.80 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 56.010, 93.930, 142.280, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.718 / 22.221 |
Other elements in 4cte:
The structure of Crystal Structure of the Catalytic Domain of the Modular Laminarinase Zglamc Mutant E142S in Complex with A Thio- Oligosaccharide also contains other interesting chemical elements:
| Chlorine | (Cl) | 2 atoms |
| Calcium | (Ca) | 2 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of the Catalytic Domain of the Modular Laminarinase Zglamc Mutant E142S in Complex with A Thio- Oligosaccharide
(pdb code 4cte). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Crystal Structure of the Catalytic Domain of the Modular Laminarinase Zglamc Mutant E142S in Complex with A Thio- Oligosaccharide, PDB code: 4cte:
In total only one binding site of Sodium was determined in the Crystal Structure of the Catalytic Domain of the Modular Laminarinase Zglamc Mutant E142S in Complex with A Thio- Oligosaccharide, PDB code: 4cte:
Sodium binding site 1 out of 1 in 4cte
Go back to
Sodium binding site 1 out
of 1 in the Crystal Structure of the Catalytic Domain of the Modular Laminarinase Zglamc Mutant E142S in Complex with A Thio- Oligosaccharide
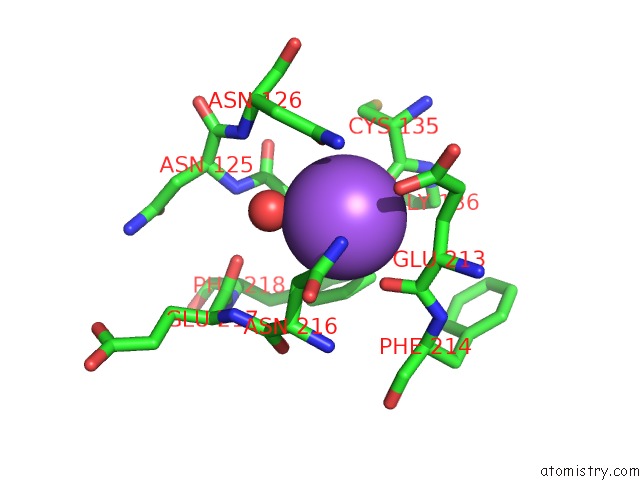
Mono view
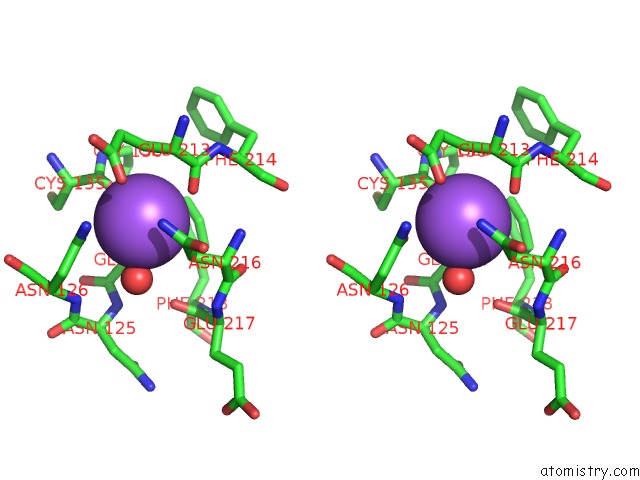
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of the Catalytic Domain of the Modular Laminarinase Zglamc Mutant E142S in Complex with A Thio- Oligosaccharide within 5.0Å range:
|
Reference:
A.Labourel,
M.Jam,
L.Legentil,
B.Sylla,
J.H.Hehemann,
V.Ferrieres,
M.Czjzek,
G.Michel.
Structural and Biochemical Characterization of the Laminarina Zglamc[GH16] From Zobellia Galactanivorans Suggests Preferred Recognition of Branched Laminarin Acta Crystallogr.,Sect.D V. 71 173 2015.
ISSN: ISSN 0907-4449
DOI: 10.1107/S139900471402450X
Page generated: Mon Oct 7 14:48:04 2024
ISSN: ISSN 0907-4449
DOI: 10.1107/S139900471402450X
Last articles
Cl in 5TQUCl in 5TQJ
Cl in 5TPI
Cl in 5TQI
Cl in 5TPU
Cl in 5TPH
Cl in 5TPX
Cl in 5TPG
Cl in 5TOW
Cl in 5TNU