Sodium »
PDB 3q11-3qpz »
3qi6 »
Sodium in PDB 3qi6: Crystal Structure of Cystathionine Gamma-Synthase Metb (Cgs) From Mycobacterium Ulcerans AGY99
Enzymatic activity of Crystal Structure of Cystathionine Gamma-Synthase Metb (Cgs) From Mycobacterium Ulcerans AGY99
All present enzymatic activity of Crystal Structure of Cystathionine Gamma-Synthase Metb (Cgs) From Mycobacterium Ulcerans AGY99:
2.5.1.48;
2.5.1.48;
Protein crystallography data
The structure of Crystal Structure of Cystathionine Gamma-Synthase Metb (Cgs) From Mycobacterium Ulcerans AGY99, PDB code: 3qi6
was solved by
Seattle Structural Genomics Center For Infectious Disease (Ssgcid),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 44.47 / 1.91 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 80.956, 106.924, 100.307, 90.00, 113.67, 90.00 |
| R / Rfree (%) | 20 / 24.1 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of Cystathionine Gamma-Synthase Metb (Cgs) From Mycobacterium Ulcerans AGY99
(pdb code 3qi6). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Crystal Structure of Cystathionine Gamma-Synthase Metb (Cgs) From Mycobacterium Ulcerans AGY99, PDB code: 3qi6:
In total only one binding site of Sodium was determined in the Crystal Structure of Cystathionine Gamma-Synthase Metb (Cgs) From Mycobacterium Ulcerans AGY99, PDB code: 3qi6:
Sodium binding site 1 out of 1 in 3qi6
Go back to
Sodium binding site 1 out
of 1 in the Crystal Structure of Cystathionine Gamma-Synthase Metb (Cgs) From Mycobacterium Ulcerans AGY99
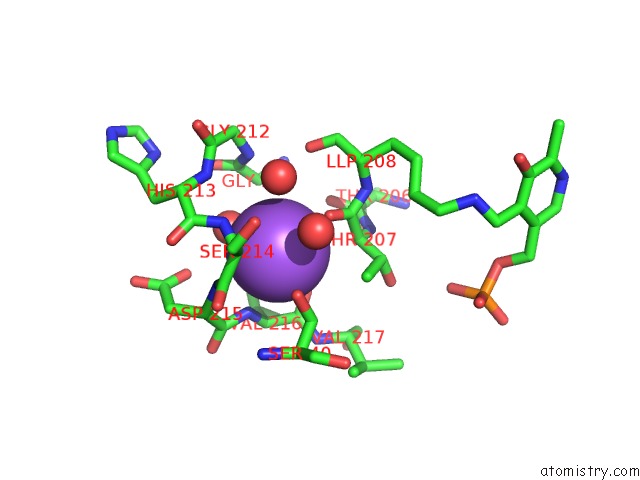
Mono view
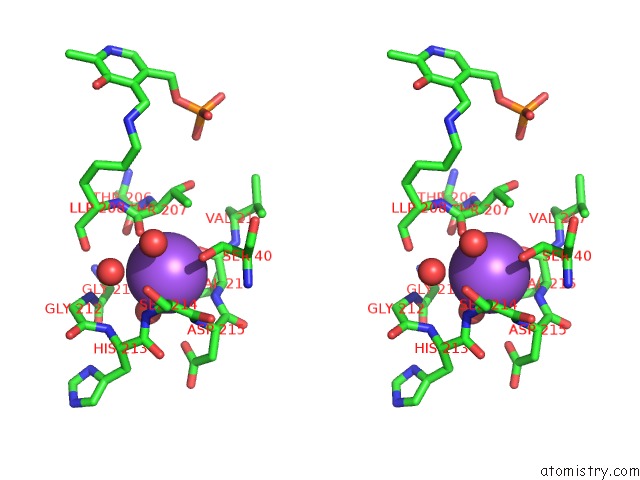
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of Cystathionine Gamma-Synthase Metb (Cgs) From Mycobacterium Ulcerans AGY99 within 5.0Å range:
|
Reference:
M.C.Clifton,
J.Abendroth,
T.E.Edwards,
D.J.Leibly,
A.K.Gillespie,
M.Ferrell,
S.H.Dieterich,
I.Exley,
B.L.Staker,
P.J.Myler,
W.C.Van Voorhis,
L.J.Stewart.
Structure of the Cystathionine [Gamma]-Synthase Metb From Mycobacterium Ulcerans Acta Crystallogr.,Sect.F V. 67 1154 2011.
ISSN: ESSN 1744-3091
PubMed: 21904066
DOI: 10.1107/S1744309111029575
Page generated: Mon Oct 7 12:33:23 2024
ISSN: ESSN 1744-3091
PubMed: 21904066
DOI: 10.1107/S1744309111029575
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO