Sodium »
PDB 3p2y-3pkh »
3p8n »
Sodium in PDB 3p8n: Crystal Structure of Hcv NS3/NS4A Protease Complexed with Bi 201335
Enzymatic activity of Crystal Structure of Hcv NS3/NS4A Protease Complexed with Bi 201335
All present enzymatic activity of Crystal Structure of Hcv NS3/NS4A Protease Complexed with Bi 201335:
3.4.21.98;
3.4.21.98;
Protein crystallography data
The structure of Crystal Structure of Hcv NS3/NS4A Protease Complexed with Bi 201335, PDB code: 3p8n
was solved by
C.T.Lemke,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.00 / 1.90 |
| Space group | P 61 |
| Cell size a, b, c (Å), α, β, γ (°) | 94.874, 94.874, 81.880, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 20.3 / 24.2 |
Other elements in 3p8n:
The structure of Crystal Structure of Hcv NS3/NS4A Protease Complexed with Bi 201335 also contains other interesting chemical elements:
| Bromine | (Br) | 2 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of Hcv NS3/NS4A Protease Complexed with Bi 201335
(pdb code 3p8n). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Crystal Structure of Hcv NS3/NS4A Protease Complexed with Bi 201335, PDB code: 3p8n:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Crystal Structure of Hcv NS3/NS4A Protease Complexed with Bi 201335, PDB code: 3p8n:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 3p8n
Go back to
Sodium binding site 1 out
of 2 in the Crystal Structure of Hcv NS3/NS4A Protease Complexed with Bi 201335
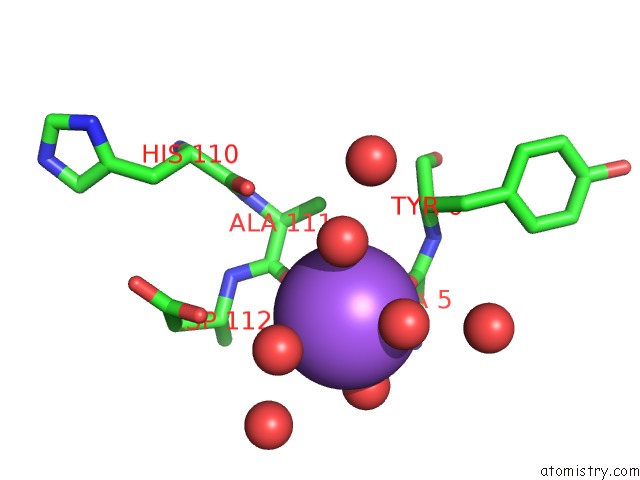
Mono view
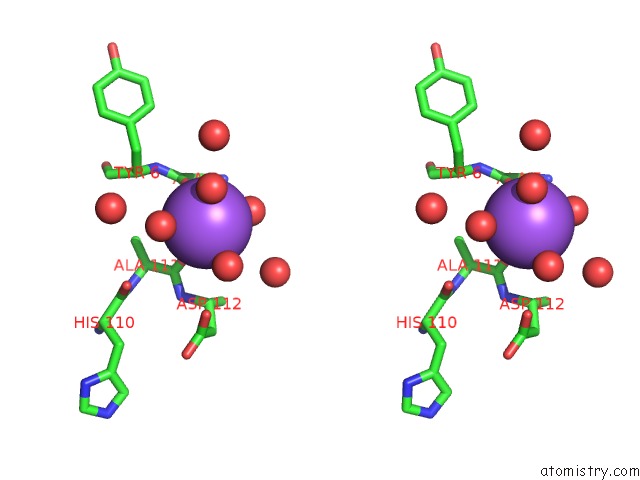
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of Hcv NS3/NS4A Protease Complexed with Bi 201335 within 5.0Å range:
|
Sodium binding site 2 out of 2 in 3p8n
Go back to
Sodium binding site 2 out
of 2 in the Crystal Structure of Hcv NS3/NS4A Protease Complexed with Bi 201335
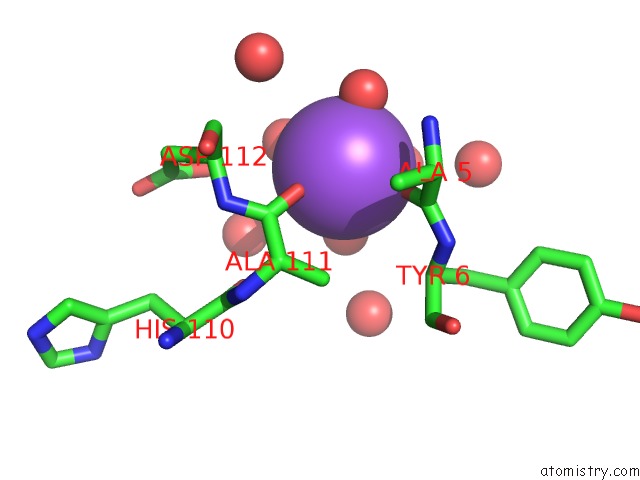
Mono view
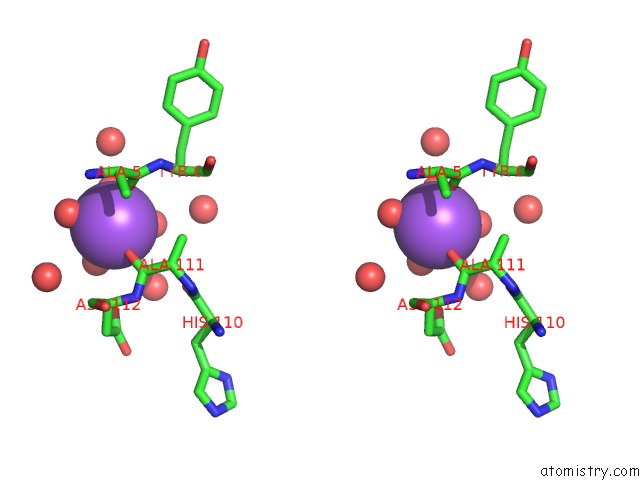
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure of Hcv NS3/NS4A Protease Complexed with Bi 201335 within 5.0Å range:
|
Reference:
C.T.Lemke,
N.Goudreau,
S.Zhao,
O.Hucke,
D.Thibeault,
M.Llinas-Brunet,
P.W.White.
Combined X-Ray, uc(Nmr), and Kinetic Analyses Reveal Uncommon Binding Characteristics of the Hepatitis C Virus NS3-NS4A Protease Inhibitor Bi 201335. J.Biol.Chem. V. 286 11434 2011.
ISSN: ISSN 0021-9258
PubMed: 21270126
DOI: 10.1074/JBC.M110.211417
Page generated: Mon Oct 7 12:11:25 2024
ISSN: ISSN 0021-9258
PubMed: 21270126
DOI: 10.1074/JBC.M110.211417
Last articles
Ca in 2YGPCa in 2YEQ
Ca in 2YG0
Ca in 2YFZ
Ca in 2YFS
Ca in 2YFU
Ca in 2YFT
Ca in 2YDP
Ca in 2YFR
Ca in 2YC4