Sodium »
PDB 3p2y-3pkh »
3p6z »
Sodium in PDB 3p6z: Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction
Enzymatic activity of Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction
All present enzymatic activity of Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction:
3.4.21.5;
3.4.21.5;
Protein crystallography data
The structure of Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction, PDB code: 3p6z
was solved by
M.A.Corral-Rodriguez,
P.E.Bock,
E.Hernandez-Carvajal,
R.Gutierrez-Gallego,
P.Fuentes-Prior,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 31.92 / 1.70 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 52.208, 62.259, 67.510, 99.38, 110.46, 92.26 |
| R / Rfree (%) | 20 / 23.9 |
Other elements in 3p6z:
The structure of Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction also contains other interesting chemical elements:
| Chlorine | (Cl) | 3 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction
(pdb code 3p6z). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 9 binding sites of Sodium where determined in the Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction, PDB code: 3p6z:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
In total 9 binding sites of Sodium where determined in the Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction, PDB code: 3p6z:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
Sodium binding site 1 out of 9 in 3p6z
Go back to
Sodium binding site 1 out
of 9 in the Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction
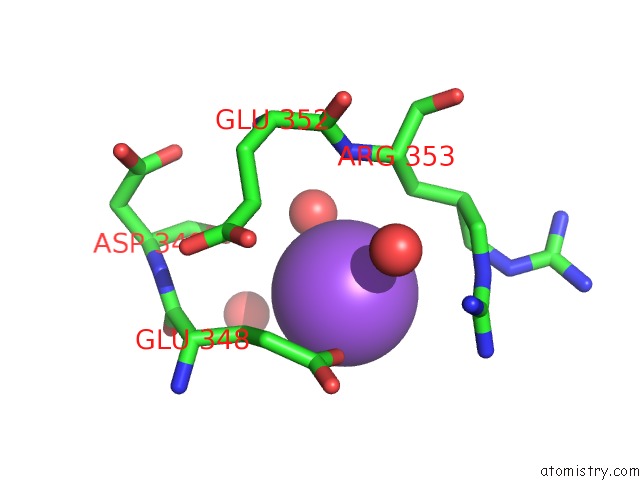
Mono view
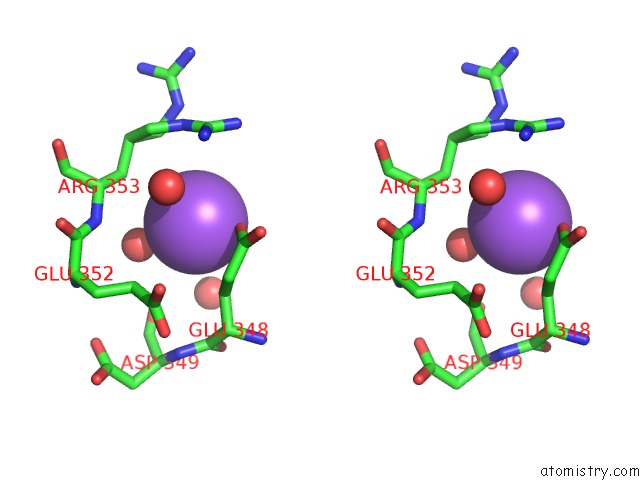
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction within 5.0Å range:
|
Sodium binding site 2 out of 9 in 3p6z
Go back to
Sodium binding site 2 out
of 9 in the Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction
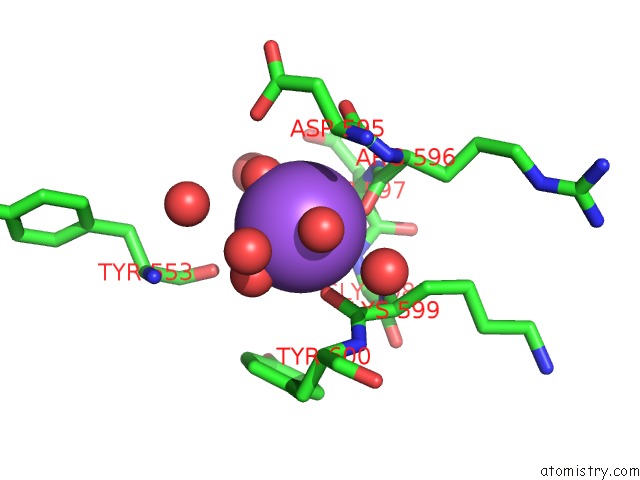
Mono view
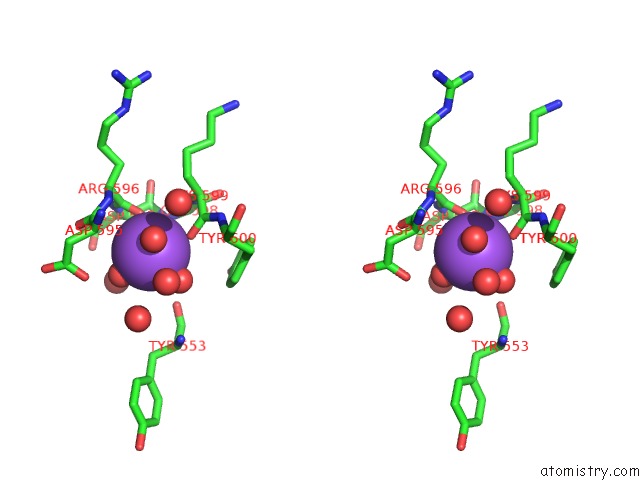
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction within 5.0Å range:
|
Sodium binding site 3 out of 9 in 3p6z
Go back to
Sodium binding site 3 out
of 9 in the Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction
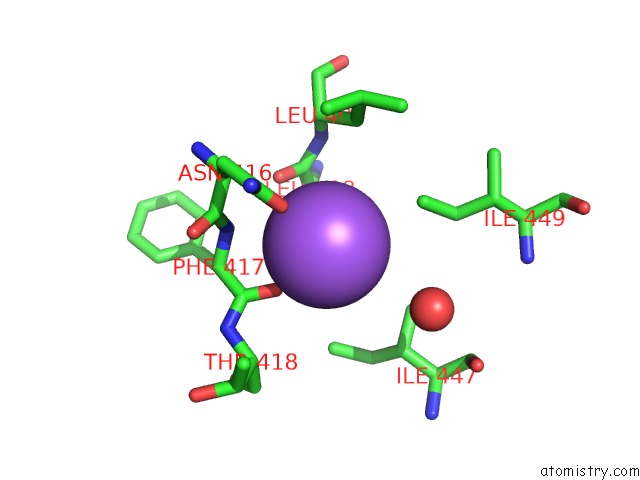
Mono view
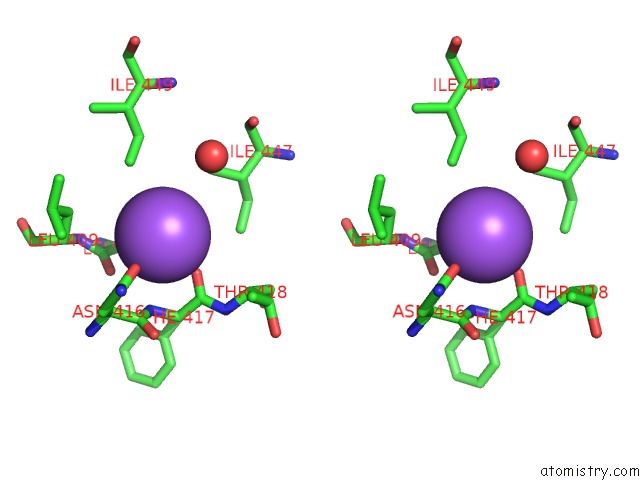
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction within 5.0Å range:
|
Sodium binding site 4 out of 9 in 3p6z
Go back to
Sodium binding site 4 out
of 9 in the Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction
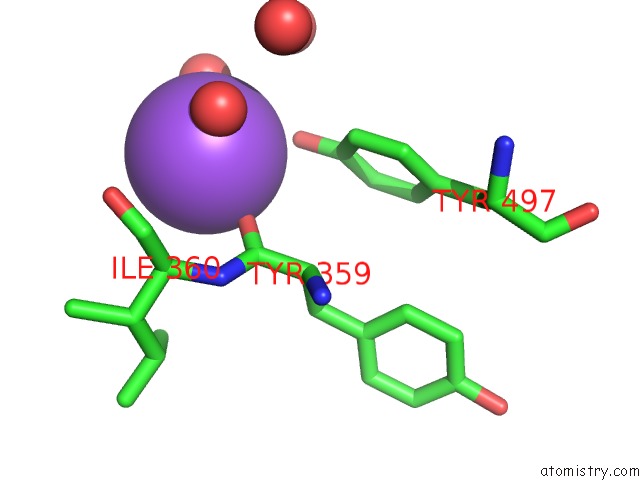
Mono view
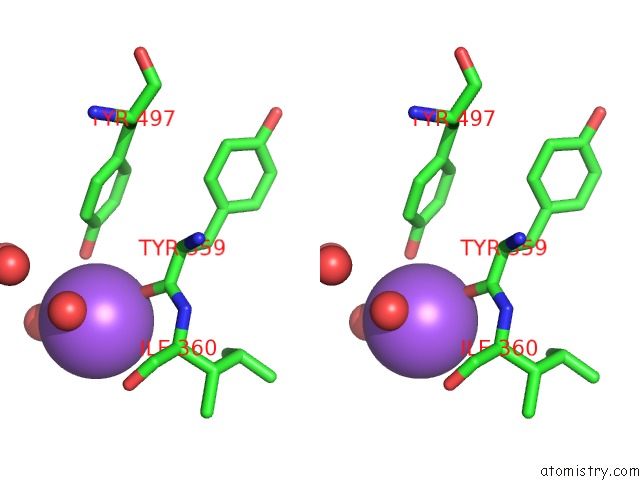
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction within 5.0Å range:
|
Sodium binding site 5 out of 9 in 3p6z
Go back to
Sodium binding site 5 out
of 9 in the Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction
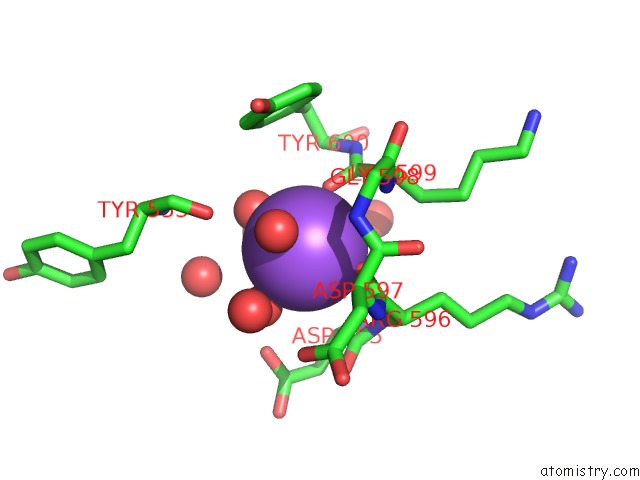
Mono view
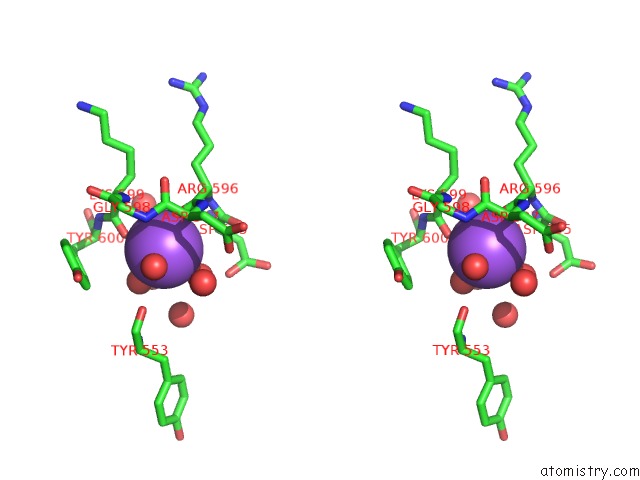
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 5 of Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction within 5.0Å range:
|
Sodium binding site 6 out of 9 in 3p6z
Go back to
Sodium binding site 6 out
of 9 in the Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction
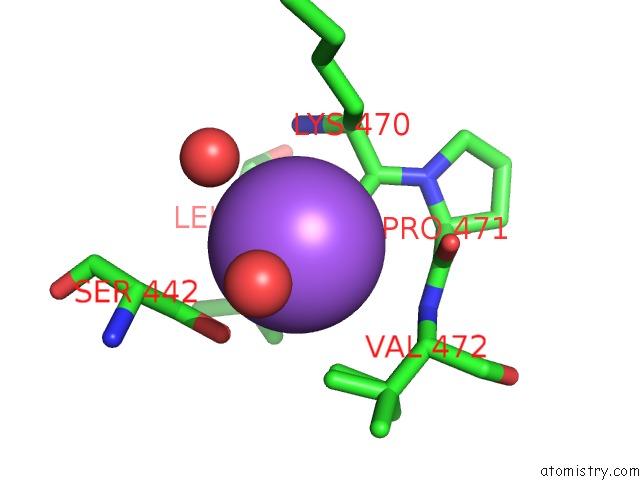
Mono view
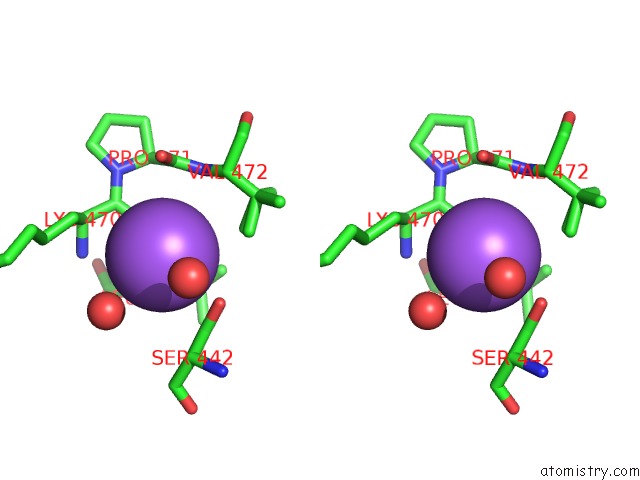
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 6 of Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction within 5.0Å range:
|
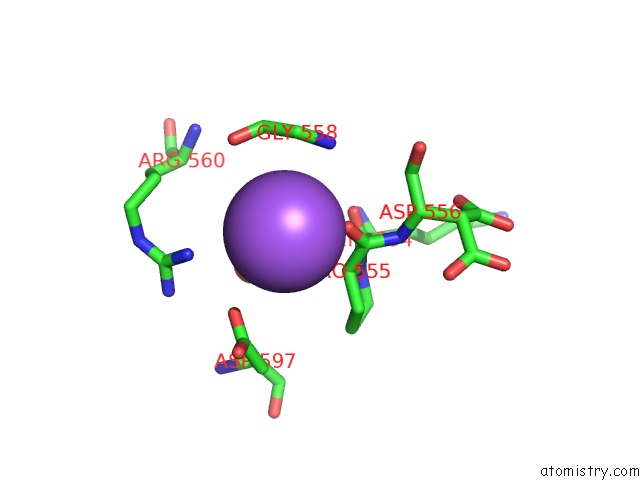
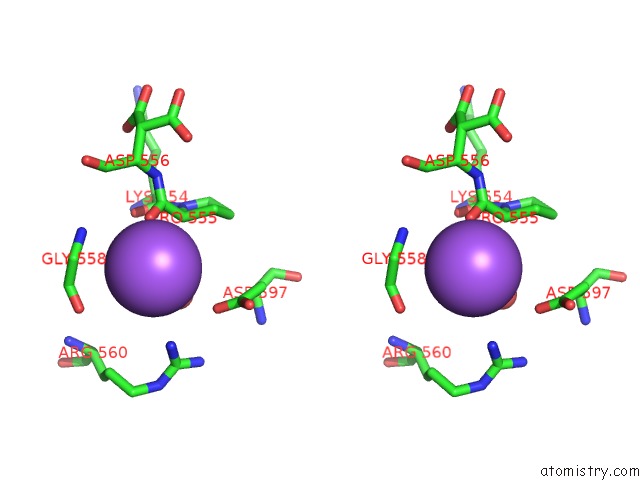
Sodium binding site 7 out of 9 in 3p6z
Go back to
Sodium binding site 7 out
of 9 in the Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 7 of Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction within 5.0Å range:
|
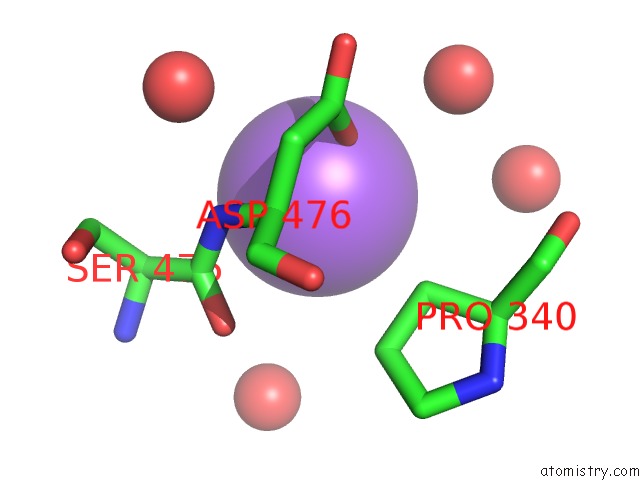
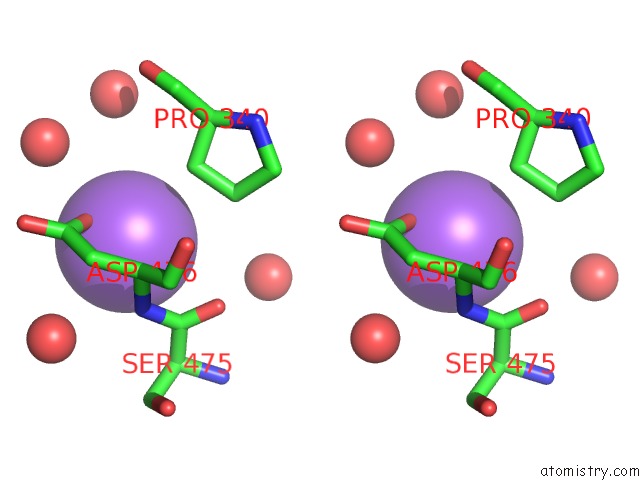
Sodium binding site 8 out of 9 in 3p6z
Go back to
Sodium binding site 8 out
of 9 in the Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 8 of Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction within 5.0Å range:
|
Sodium binding site 9 out of 9 in 3p6z
Go back to
Sodium binding site 9 out
of 9 in the Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction
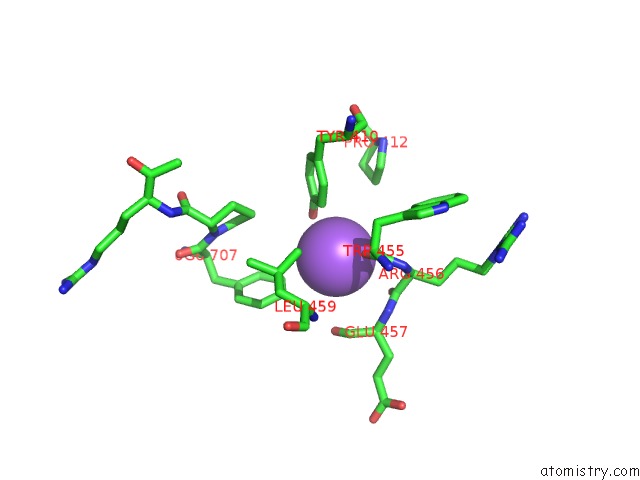
Mono view
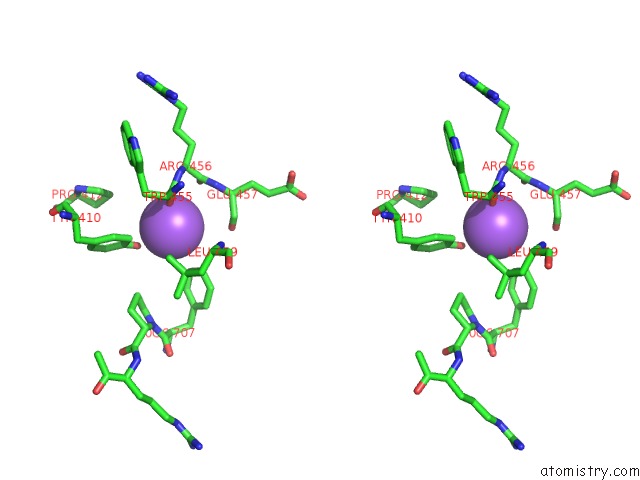
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 9 of Structural Basis of Thrombin Mediated Factor V Activation: Essential Role of the Hirudin-Like Sequence GLU666-GLU672 For Processing at the Heavy Chain-B Domain Junction within 5.0Å range:
|
Reference:
M.A.Corral-Rodriguez,
P.E.Bock,
E.Hernandez-Carvajal,
R.Gutierrez-Gallego,
P.Fuentes-Prior.
Structural Basis of Thrombin-Mediated Factor V Activation: the GLU666-GLU672 Sequence Is Critical For Processing at the Heavy Chain-B Domain Junction. Blood V. 117 7164 2011.
ISSN: ISSN 0006-4971
PubMed: 21555742
DOI: 10.1182/BLOOD-2010-10-315309
Page generated: Mon Oct 7 12:11:25 2024
ISSN: ISSN 0006-4971
PubMed: 21555742
DOI: 10.1182/BLOOD-2010-10-315309
Last articles
Cl in 5G0BCl in 5FZO
Cl in 5FZL
Cl in 5FZK
Cl in 5FZM
Cl in 5FZI
Cl in 5FZH
Cl in 5FZG
Cl in 5FZF
Cl in 5FZE