Sodium »
PDB 3nte-3off »
3nvd »
Sodium in PDB 3nvd: Structure of Ybbd in Complex with Pugnac
Enzymatic activity of Structure of Ybbd in Complex with Pugnac
All present enzymatic activity of Structure of Ybbd in Complex with Pugnac:
3.2.1.52;
3.2.1.52;
Protein crystallography data
The structure of Structure of Ybbd in Complex with Pugnac, PDB code: 3nvd
was solved by
K.Diederichs,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.55 / 1.84 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.473, 73.206, 83.788, 79.83, 69.45, 88.22 |
| R / Rfree (%) | 18.7 / 24.6 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Structure of Ybbd in Complex with Pugnac
(pdb code 3nvd). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Structure of Ybbd in Complex with Pugnac, PDB code: 3nvd:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Structure of Ybbd in Complex with Pugnac, PDB code: 3nvd:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 3nvd
Go back to
Sodium binding site 1 out
of 2 in the Structure of Ybbd in Complex with Pugnac
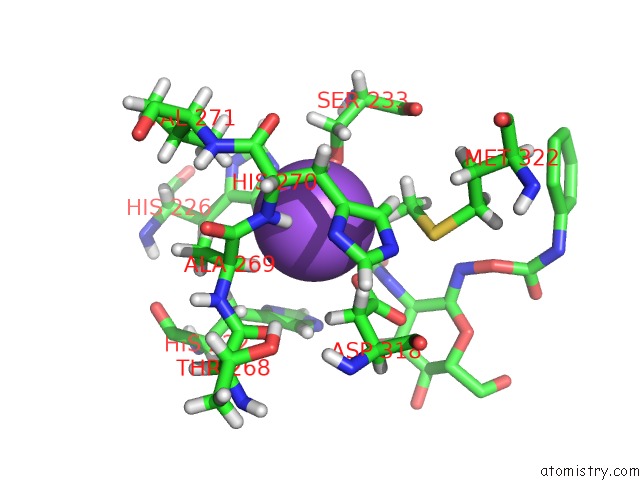
Mono view
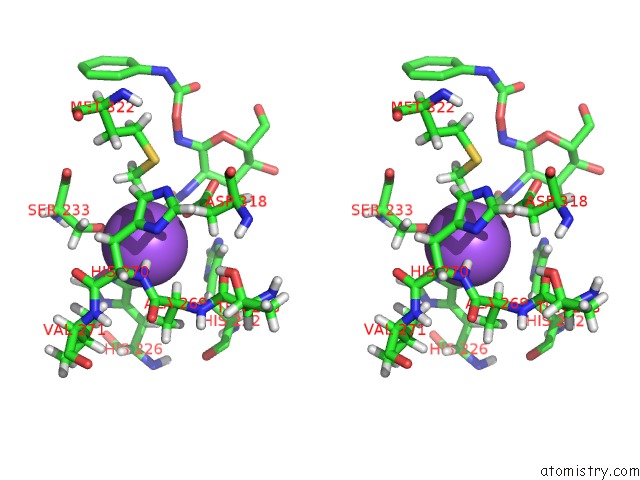
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Structure of Ybbd in Complex with Pugnac within 5.0Å range:
|
Sodium binding site 2 out of 2 in 3nvd
Go back to
Sodium binding site 2 out
of 2 in the Structure of Ybbd in Complex with Pugnac
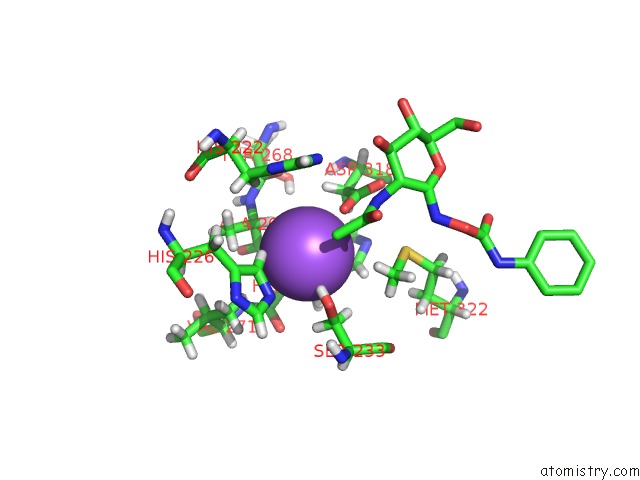
Mono view
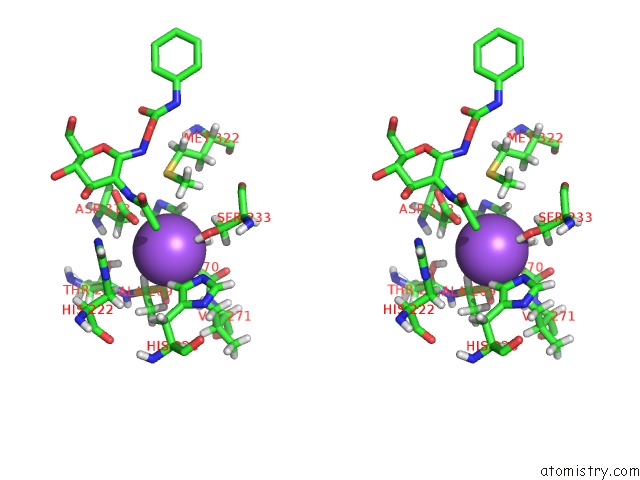
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Structure of Ybbd in Complex with Pugnac within 5.0Å range:
|
Reference:
S.Litzinger,
S.Fischer,
P.Polzer,
K.Diederichs,
W.Welte,
C.Mayer.
Structural and Kinetic Analysis of Bacillus Subtilis N-Acetylglucosaminidase Reveals A Unique Asp-His Dyad Mechanism J.Biol.Chem. V. 285 35675 2010.
ISSN: ISSN 0021-9258
PubMed: 20826810
DOI: 10.1074/JBC.M110.131037
Page generated: Mon Oct 7 11:55:00 2024
ISSN: ISSN 0021-9258
PubMed: 20826810
DOI: 10.1074/JBC.M110.131037
Last articles
F in 7M2FF in 7M0Y
F in 7M0Z
F in 7M0M
F in 7M0V
F in 7M0W
F in 7M0U
F in 7M0N
F in 7M0T
F in 7M00