Sodium »
PDB 3lbg-3m9y »
3lk9 »
Sodium in PDB 3lk9: Dna Polymerase Beta with A Gapped Dna Substrate and Dtmp(CF2)P(CF2)P
Enzymatic activity of Dna Polymerase Beta with A Gapped Dna Substrate and Dtmp(CF2)P(CF2)P
All present enzymatic activity of Dna Polymerase Beta with A Gapped Dna Substrate and Dtmp(CF2)P(CF2)P:
2.7.7.7;
2.7.7.7;
Protein crystallography data
The structure of Dna Polymerase Beta with A Gapped Dna Substrate and Dtmp(CF2)P(CF2)P, PDB code: 3lk9
was solved by
M.Zibinsky,
G.K.Surya Prakash,
T.G.Upton,
B.A.Kashemirov,
C.E.Mckenna,
K.Oertell,
M.F.Goodman,
V.K.Batra,
L.C.Pedersen,
W.A.Beard,
S.H.Wilson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.28 / 2.50 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 49.960, 79.800, 55.050, 90.00, 108.46, 90.00 |
| R / Rfree (%) | 19.8 / 28.1 |
Other elements in 3lk9:
The structure of Dna Polymerase Beta with A Gapped Dna Substrate and Dtmp(CF2)P(CF2)P also contains other interesting chemical elements:
| Fluorine | (F) | 4 atoms |
| Magnesium | (Mg) | 2 atoms |
| Chlorine | (Cl) | 3 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Dna Polymerase Beta with A Gapped Dna Substrate and Dtmp(CF2)P(CF2)P
(pdb code 3lk9). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Dna Polymerase Beta with A Gapped Dna Substrate and Dtmp(CF2)P(CF2)P, PDB code: 3lk9:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Dna Polymerase Beta with A Gapped Dna Substrate and Dtmp(CF2)P(CF2)P, PDB code: 3lk9:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 3lk9
Go back to
Sodium binding site 1 out
of 2 in the Dna Polymerase Beta with A Gapped Dna Substrate and Dtmp(CF2)P(CF2)P

Mono view
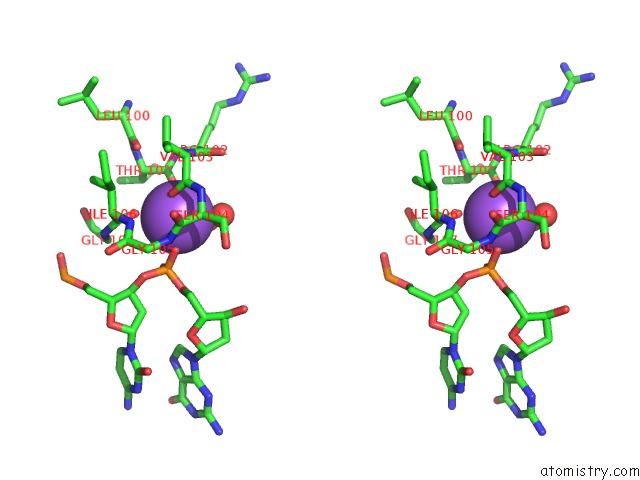
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Dna Polymerase Beta with A Gapped Dna Substrate and Dtmp(CF2)P(CF2)P within 5.0Å range:
|
Sodium binding site 2 out of 2 in 3lk9
Go back to
Sodium binding site 2 out
of 2 in the Dna Polymerase Beta with A Gapped Dna Substrate and Dtmp(CF2)P(CF2)P

Mono view
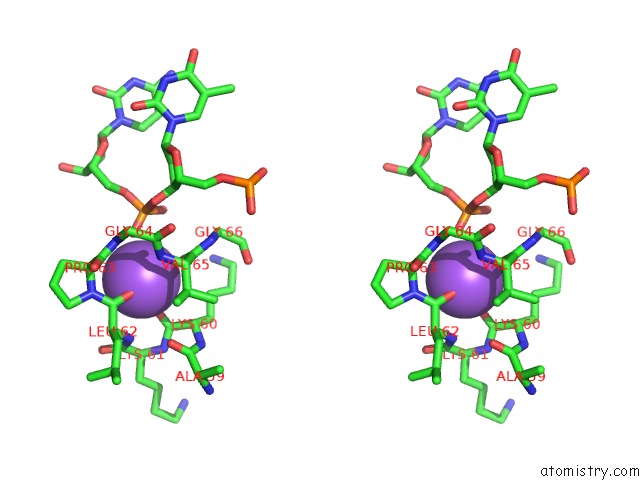
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Dna Polymerase Beta with A Gapped Dna Substrate and Dtmp(CF2)P(CF2)P within 5.0Å range:
|
Reference:
G.K.Surya Prakash,
M.Zibinsky,
T.G.Upton,
B.A.Kashemirov,
C.E.Mckenna,
K.Oertell,
M.F.Goodman,
V.K.Batra,
L.C.Pedersen,
W.A.Beard,
D.D.Shock,
S.H.Wilson,
G.A.Olah.
Synthesis and Biological Evaluation of Fluorinated Deoxynucleotide Analogs Based on Bis-(Difluoromethylene)Triphosphoric Acid. Proc.Natl.Acad.Sci.Usa V. 107 15693 2010.
ISSN: ISSN 0027-8424
PubMed: 20724659
DOI: 10.1073/PNAS.1007430107
Page generated: Mon Oct 7 11:22:20 2024
ISSN: ISSN 0027-8424
PubMed: 20724659
DOI: 10.1073/PNAS.1007430107
Last articles
Cl in 5J2BCl in 5J2A
Cl in 5IXL
Cl in 5J0Z
Cl in 5J1T
Cl in 5J1S
Cl in 5IXY
Cl in 5IXE
Cl in 5IXS
Cl in 5IVT