Sodium »
PDB 3epz-3f9l »
3f7k »
Sodium in PDB 3f7k: X-Ray Crystal Structure of An Alvinella Pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex
Enzymatic activity of X-Ray Crystal Structure of An Alvinella Pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex
All present enzymatic activity of X-Ray Crystal Structure of An Alvinella Pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex:
1.15.1.1;
1.15.1.1;
Protein crystallography data
The structure of X-Ray Crystal Structure of An Alvinella Pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex, PDB code: 3f7k
was solved by
D.S.Shin,
M.Didonato,
D.P.Barondeau,
E.D.Getzoff,
J.A.Tainer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.00 / 1.35 |
| Space group | P 61 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 62.563, 62.563, 163.745, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 12.8 / 17.6 |
Other elements in 3f7k:
The structure of X-Ray Crystal Structure of An Alvinella Pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex also contains other interesting chemical elements:
| Copper | (Cu) | 2 atoms |
| Zinc | (Zn) | 1 atom |
Sodium Binding Sites:
The binding sites of Sodium atom in the X-Ray Crystal Structure of An Alvinella Pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex
(pdb code 3f7k). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the X-Ray Crystal Structure of An Alvinella Pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex, PDB code: 3f7k:
In total only one binding site of Sodium was determined in the X-Ray Crystal Structure of An Alvinella Pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex, PDB code: 3f7k:
Sodium binding site 1 out of 1 in 3f7k
Go back to
Sodium binding site 1 out
of 1 in the X-Ray Crystal Structure of An Alvinella Pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex
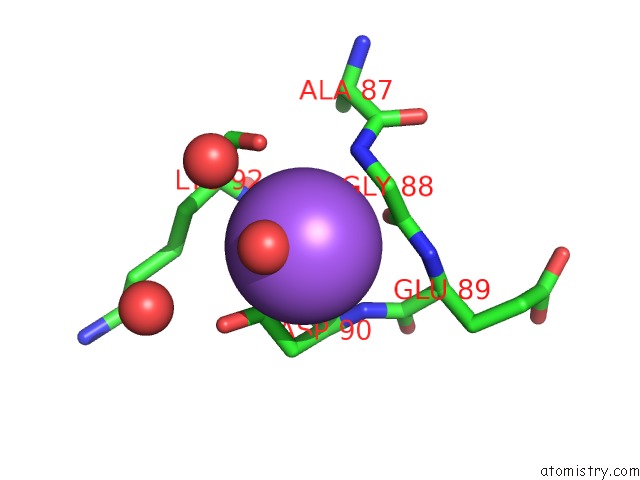
Mono view
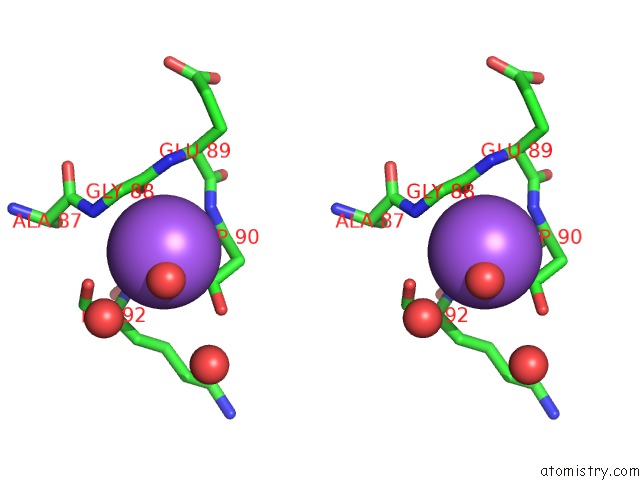
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of X-Ray Crystal Structure of An Alvinella Pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex within 5.0Å range:
|
Reference:
D.S.Shin,
M.Didonato,
D.P.Barondeau,
G.L.Hura,
C.Hitomi,
J.A.Berglund,
E.D.Getzoff,
S.C.Cary,
J.A.Tainer.
Superoxide Dismutase From the Eukaryotic Thermophile Alvinella Pompejana: Structures, Stability, Mechanism, and Insights Into Amyotrophic Lateral Sclerosis. J.Mol.Biol. V. 385 1534 2009.
ISSN: ISSN 0022-2836
PubMed: 19063897
DOI: 10.1016/J.JMB.2008.11.031
Page generated: Mon Oct 7 08:59:05 2024
ISSN: ISSN 0022-2836
PubMed: 19063897
DOI: 10.1016/J.JMB.2008.11.031
Last articles
Cl in 5FMACl in 5FLP
Cl in 5FLF
Cl in 5FLS
Cl in 5FLO
Cl in 5FLD
Cl in 5FKN
Cl in 5FKO
Cl in 5FL3
Cl in 5FKK