Sodium »
PDB 3epz-3f9l »
3f48 »
Sodium in PDB 3f48: Crystal Structure of Leut Bound to L-Alanine and Sodium
Protein crystallography data
The structure of Crystal Structure of Leut Bound to L-Alanine and Sodium, PDB code: 3f48
was solved by
S.K.Singh,
C.L.Piscitelli,
A.Yamashita,
E.Gouaux,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.68 / 1.90 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 89.400, 87.200, 81.200, 90.00, 95.90, 90.00 |
| R / Rfree (%) | 21.1 / 22.9 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of Leut Bound to L-Alanine and Sodium
(pdb code 3f48). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Crystal Structure of Leut Bound to L-Alanine and Sodium, PDB code: 3f48:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Crystal Structure of Leut Bound to L-Alanine and Sodium, PDB code: 3f48:
Jump to Sodium binding site number: 1; 2;
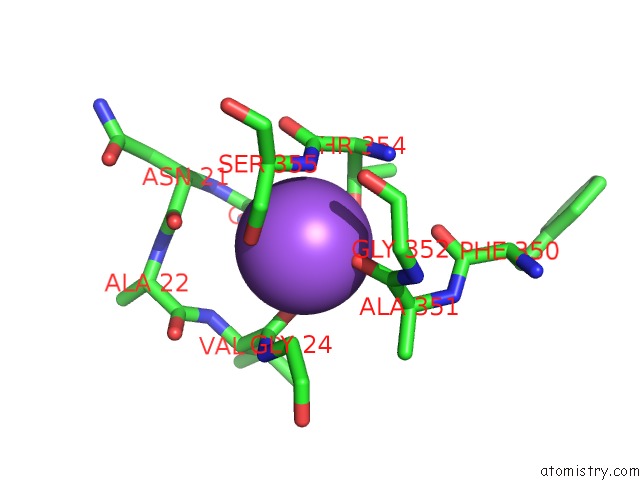
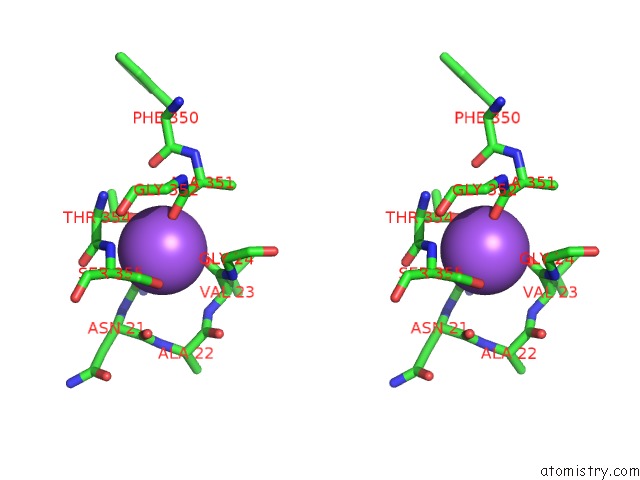
Sodium binding site 1 out of 2 in 3f48
Go back to
Sodium binding site 1 out
of 2 in the Crystal Structure of Leut Bound to L-Alanine and Sodium
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of Leut Bound to L-Alanine and Sodium within 5.0Å range:
|
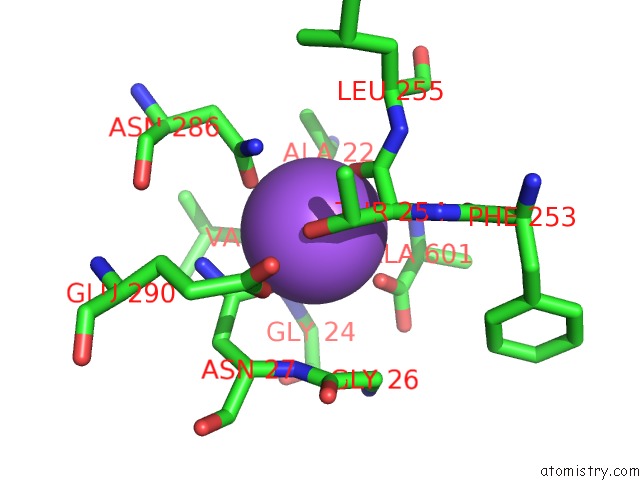
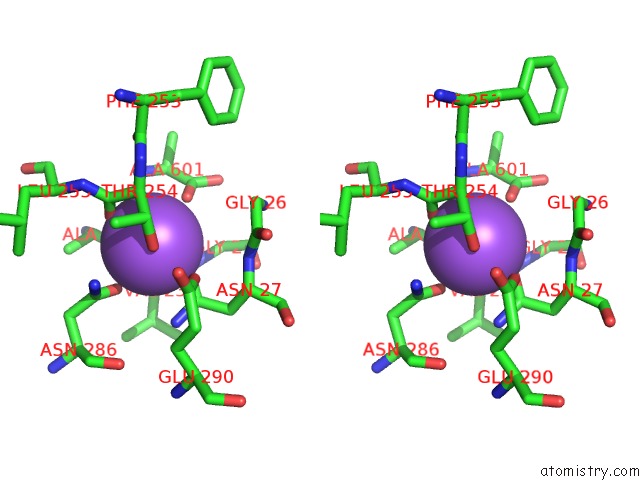
Sodium binding site 2 out of 2 in 3f48
Go back to
Sodium binding site 2 out
of 2 in the Crystal Structure of Leut Bound to L-Alanine and Sodium
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure of Leut Bound to L-Alanine and Sodium within 5.0Å range:
|
Reference:
S.K.Singh,
C.L.Piscitelli,
A.Yamashita,
E.Gouaux.
A Competitive Inhibitor Traps Leut in An Open-to-Out Conformation. Science V. 322 1655 2008.
ISSN: ISSN 0036-8075
PubMed: 19074341
DOI: 10.1126/SCIENCE.1166777
Page generated: Mon Oct 7 08:56:39 2024
ISSN: ISSN 0036-8075
PubMed: 19074341
DOI: 10.1126/SCIENCE.1166777
Last articles
Cl in 7UODCl in 7UN0
Cl in 7UOQ
Cl in 7UOC
Cl in 7UNO
Cl in 7UNN
Cl in 7UN2
Cl in 7UMP
Cl in 7UMG
Cl in 7UKT