Sodium »
PDB 2vrr-2wcp »
2vu1 »
Sodium in PDB 2vu1: Biosynthetic Thiolase From Z. Ramigera. Complex of with O-Pantheteine- 11-Pivalate.
Enzymatic activity of Biosynthetic Thiolase From Z. Ramigera. Complex of with O-Pantheteine- 11-Pivalate.
All present enzymatic activity of Biosynthetic Thiolase From Z. Ramigera. Complex of with O-Pantheteine- 11-Pivalate.:
2.3.1.9;
2.3.1.9;
Protein crystallography data
The structure of Biosynthetic Thiolase From Z. Ramigera. Complex of with O-Pantheteine- 11-Pivalate., PDB code: 2vu1
was solved by
P.Kursula,
W.Schmitz,
R.K.Wierenga,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 1.51 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 84.298, 78.737, 148.339, 90.00, 92.93, 90.00 |
| R / Rfree (%) | 21.5 / 24.7 |
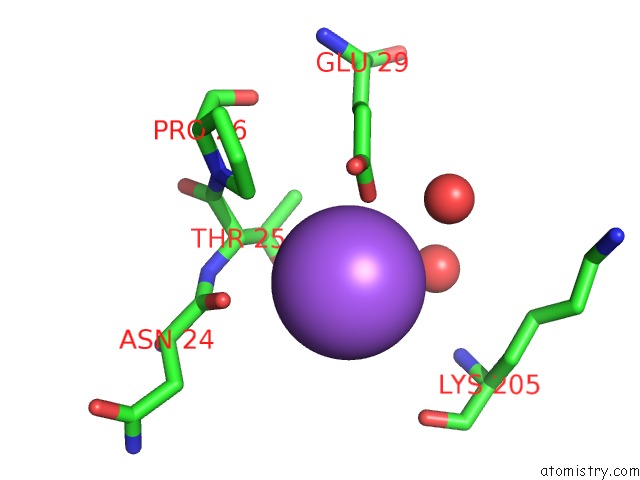
Sodium Binding Sites:
The binding sites of Sodium atom in the Biosynthetic Thiolase From Z. Ramigera. Complex of with O-Pantheteine- 11-Pivalate.
(pdb code 2vu1). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Biosynthetic Thiolase From Z. Ramigera. Complex of with O-Pantheteine- 11-Pivalate., PDB code: 2vu1:
In total only one binding site of Sodium was determined in the Biosynthetic Thiolase From Z. Ramigera. Complex of with O-Pantheteine- 11-Pivalate., PDB code: 2vu1:
Sodium binding site 1 out of 1 in 2vu1
Go back to
Sodium binding site 1 out
of 1 in the Biosynthetic Thiolase From Z. Ramigera. Complex of with O-Pantheteine- 11-Pivalate.
Mono view
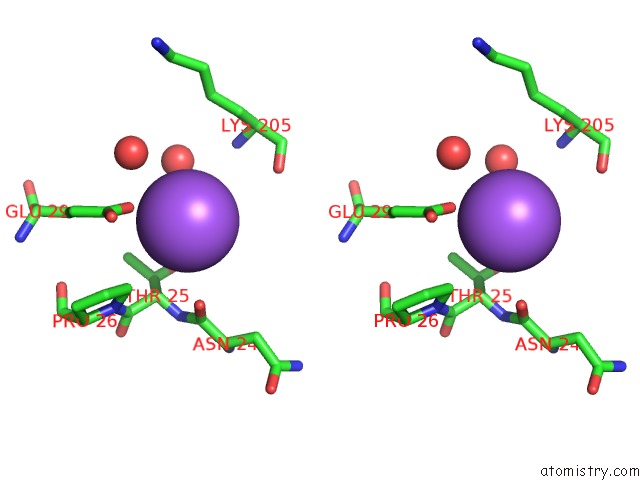
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Biosynthetic Thiolase From Z. Ramigera. Complex of with O-Pantheteine- 11-Pivalate. within 5.0Å range:
|
Reference:
G.Merilainen,
W.Schmitz,
R.K.Wierenga,
P.Kursula.
The Sulfur Atoms of the Substrate Coa and the Catalytic Cysteine Are Required For A Productive Mode of Substrate Binding in Bacterial Biosynthetic Thiolase, A Thioester-Dependent Enzyme. Febs J. V. 275 6136 2008.
ISSN: ISSN 1742-464X
PubMed: 19016856
DOI: 10.1111/J.1742-4658.2008.06737.X
Page generated: Mon Oct 7 04:31:15 2024
ISSN: ISSN 1742-464X
PubMed: 19016856
DOI: 10.1111/J.1742-4658.2008.06737.X
Last articles
Ca in 5MHCCa in 5MFL
Ca in 5MFN
Ca in 5MFH
Ca in 5MFE
Ca in 5MFG
Ca in 5MFA
Ca in 5MF4
Ca in 5MEY
Ca in 5MB1