Sodium »
PDB 2ply-2qd6 »
2qd6 »
Sodium in PDB 2qd6: Hiv-1 Protease Mutant I50V with Potent Antiviral Inhibitor Grl-98065
Enzymatic activity of Hiv-1 Protease Mutant I50V with Potent Antiviral Inhibitor Grl-98065
All present enzymatic activity of Hiv-1 Protease Mutant I50V with Potent Antiviral Inhibitor Grl-98065:
3.4.23.16;
3.4.23.16;
Protein crystallography data
The structure of Hiv-1 Protease Mutant I50V with Potent Antiviral Inhibitor Grl-98065, PDB code: 2qd6
was solved by
Y.F.Wang,
Y.Tie,
P.I.Boross,
J.Tozser,
A.K.Ghosh,
R.W.Harrison,
I.T.Weber,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 10.00 / 1.28 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.294, 86.228, 46.006, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.5 / 20.2 |
Other elements in 2qd6:
The structure of Hiv-1 Protease Mutant I50V with Potent Antiviral Inhibitor Grl-98065 also contains other interesting chemical elements:
| Chlorine | (Cl) | 2 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Hiv-1 Protease Mutant I50V with Potent Antiviral Inhibitor Grl-98065
(pdb code 2qd6). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Hiv-1 Protease Mutant I50V with Potent Antiviral Inhibitor Grl-98065, PDB code: 2qd6:
In total only one binding site of Sodium was determined in the Hiv-1 Protease Mutant I50V with Potent Antiviral Inhibitor Grl-98065, PDB code: 2qd6:
Sodium binding site 1 out of 1 in 2qd6
Go back to
Sodium binding site 1 out
of 1 in the Hiv-1 Protease Mutant I50V with Potent Antiviral Inhibitor Grl-98065
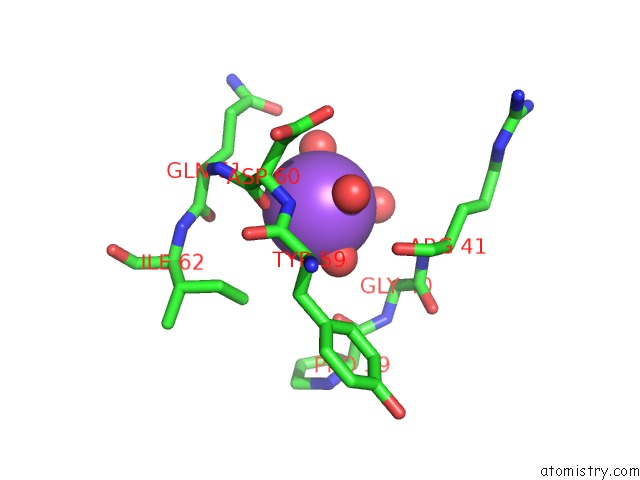
Mono view
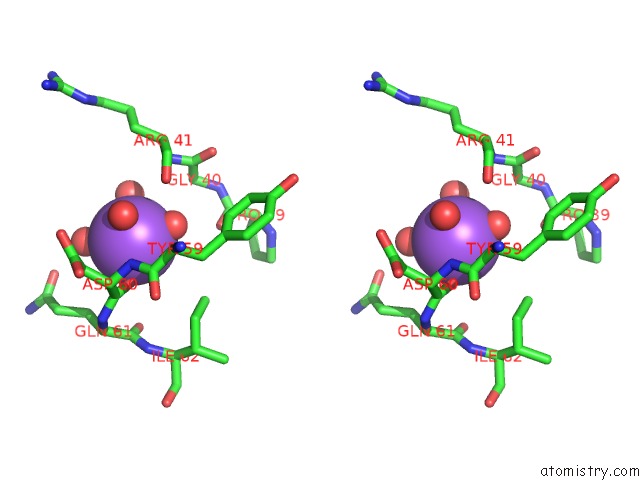
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Hiv-1 Protease Mutant I50V with Potent Antiviral Inhibitor Grl-98065 within 5.0Å range:
|
Reference:
Y.F.Wang,
Y.Tie,
P.I.Boross,
J.Tozser,
A.K.Ghosh,
R.W.Harrison,
I.T.Weber.
Potent New Antiviral Compound Shows Similar Inhibition and Structural Interactions with Drug Resistant Mutants and Wild Type Hiv-1 Protease. J.Med.Chem. V. 50 4509 2007.
ISSN: ISSN 0022-2623
PubMed: 17696515
DOI: 10.1021/JM070482Q
Page generated: Mon Oct 7 03:49:29 2024
ISSN: ISSN 0022-2623
PubMed: 17696515
DOI: 10.1021/JM070482Q
Last articles
F in 7NWEF in 7NWK
F in 7NWA
F in 7NVN
F in 7NVM
F in 7NW2
F in 7NW0
F in 7NVL
F in 7NVX
F in 7NVV