Sodium »
PDB 2p0o-2plo »
2p2t »
Sodium in PDB 2p2t: Crystal Structure of Dynein Light Chain LC8 Bound to Residues 123-138 of Intermediate Chain IC74
Protein crystallography data
The structure of Crystal Structure of Dynein Light Chain LC8 Bound to Residues 123-138 of Intermediate Chain IC74, PDB code: 2p2t
was solved by
G.Benison,
P.A.Karplus,
E.Barbar,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 22.18 / 3.00 |
| Space group | P 61 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 44.356, 44.356, 204.466, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 21 / 29.6 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of Dynein Light Chain LC8 Bound to Residues 123-138 of Intermediate Chain IC74
(pdb code 2p2t). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Crystal Structure of Dynein Light Chain LC8 Bound to Residues 123-138 of Intermediate Chain IC74, PDB code: 2p2t:
In total only one binding site of Sodium was determined in the Crystal Structure of Dynein Light Chain LC8 Bound to Residues 123-138 of Intermediate Chain IC74, PDB code: 2p2t:
Sodium binding site 1 out of 1 in 2p2t
Go back to
Sodium binding site 1 out
of 1 in the Crystal Structure of Dynein Light Chain LC8 Bound to Residues 123-138 of Intermediate Chain IC74
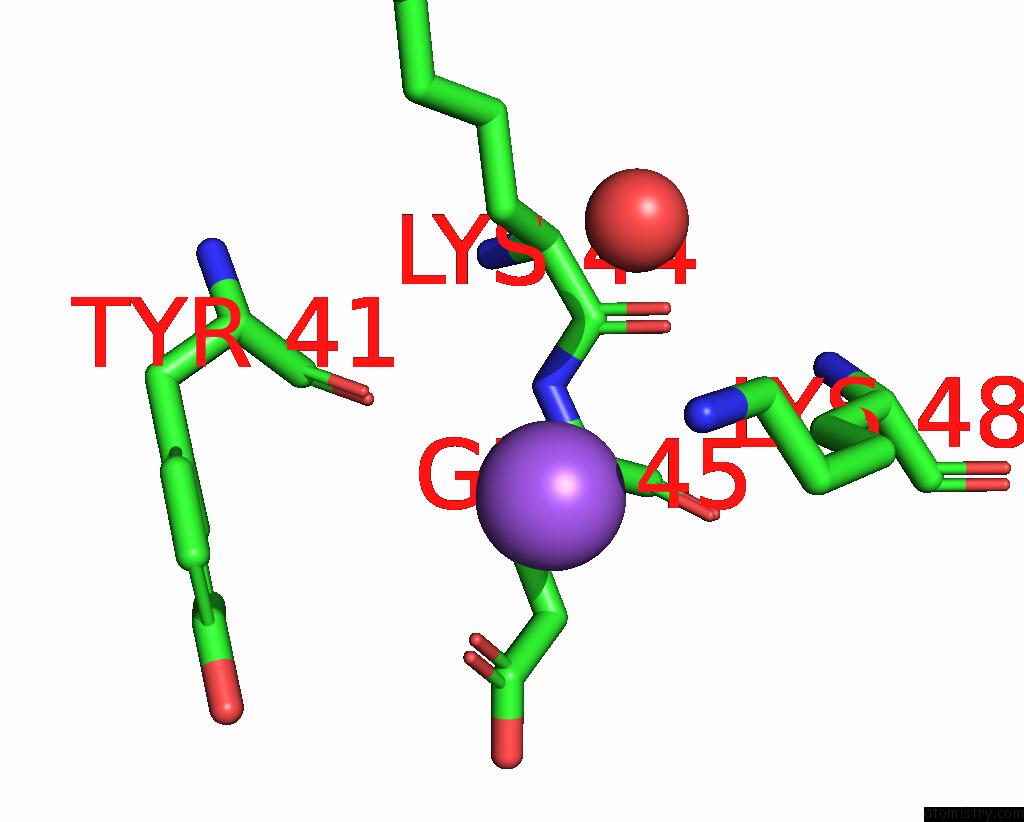
Mono view
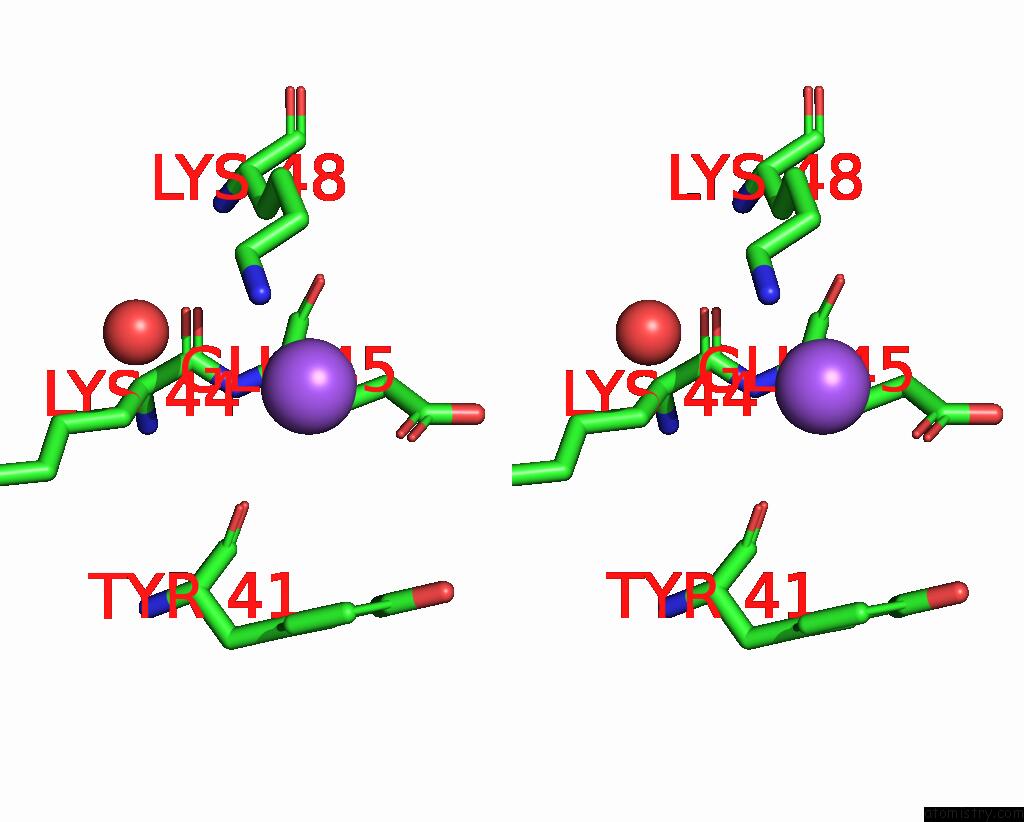
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of Dynein Light Chain LC8 Bound to Residues 123-138 of Intermediate Chain IC74 within 5.0Å range:
|
Reference:
G.Benison,
P.A.Karplus,
E.Barbar.
Structure and Dynamics of LC8 Complexes with Kxtqt-Motif Peptides: Swallow and Dynein Intermediate Chain Compete For A Common Site. J.Mol.Biol. V. 371 457 2007.
ISSN: ISSN 0022-2836
PubMed: 17570393
DOI: 10.1016/J.JMB.2007.05.046
Page generated: Mon Oct 7 03:37:52 2024
ISSN: ISSN 0022-2836
PubMed: 17570393
DOI: 10.1016/J.JMB.2007.05.046
Last articles
Ca in 2XMDCa in 2XJV
Ca in 2XJU
Ca in 2XJT
Ca in 2XJO
Ca in 2XJM
Ca in 2XJS
Ca in 2XJR
Ca in 2XJN
Ca in 2XJP