Sodium »
PDB 2htx-2isp »
2hzl »
Sodium in PDB 2hzl: Crystal Structures of A Sodium-Alpha-Keto Acid Binding Subunit From A Trap Transporter in Its Closed Forms
Protein crystallography data
The structure of Crystal Structures of A Sodium-Alpha-Keto Acid Binding Subunit From A Trap Transporter in Its Closed Forms, PDB code: 2hzl
was solved by
S.Gonin,
P.Arnoux,
B.Pierru,
B.Alonso,
M.Sabaty,
D.Pignol,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 39.01 / 1.40 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 118.021, 78.057, 95.139, 90.00, 124.98, 90.00 |
| R / Rfree (%) | 17.3 / 18.4 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structures of A Sodium-Alpha-Keto Acid Binding Subunit From A Trap Transporter in Its Closed Forms
(pdb code 2hzl). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Crystal Structures of A Sodium-Alpha-Keto Acid Binding Subunit From A Trap Transporter in Its Closed Forms, PDB code: 2hzl:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Crystal Structures of A Sodium-Alpha-Keto Acid Binding Subunit From A Trap Transporter in Its Closed Forms, PDB code: 2hzl:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 2hzl
Go back to
Sodium binding site 1 out
of 2 in the Crystal Structures of A Sodium-Alpha-Keto Acid Binding Subunit From A Trap Transporter in Its Closed Forms
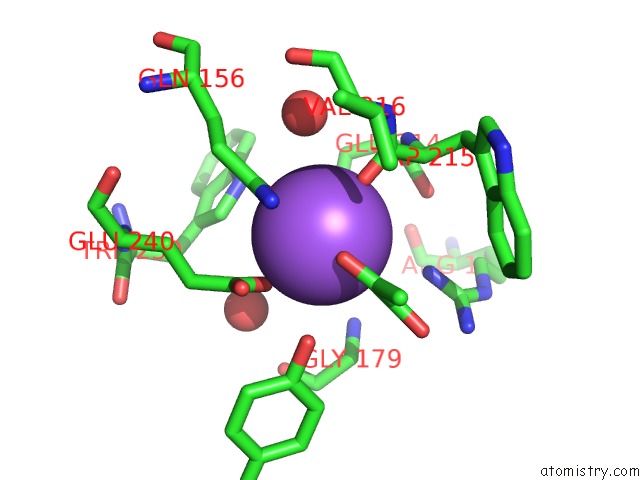
Mono view
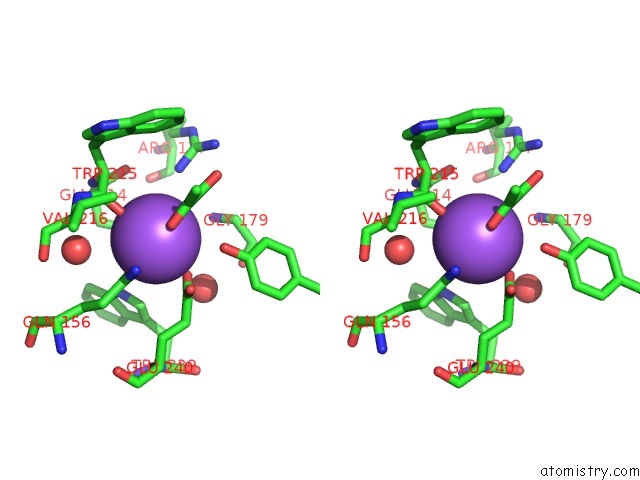
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structures of A Sodium-Alpha-Keto Acid Binding Subunit From A Trap Transporter in Its Closed Forms within 5.0Å range:
|
Sodium binding site 2 out of 2 in 2hzl
Go back to
Sodium binding site 2 out
of 2 in the Crystal Structures of A Sodium-Alpha-Keto Acid Binding Subunit From A Trap Transporter in Its Closed Forms
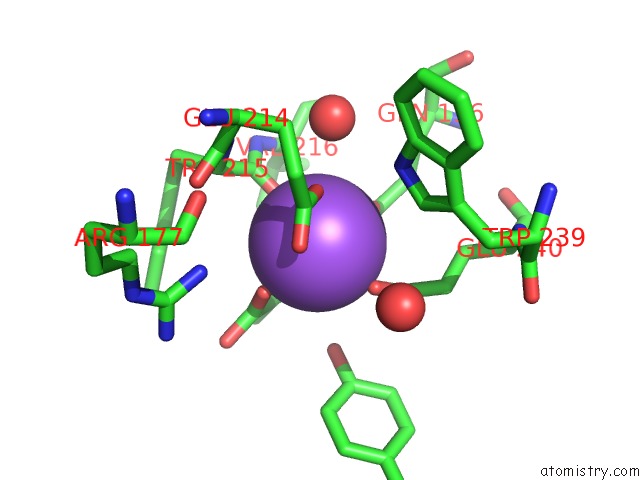
Mono view
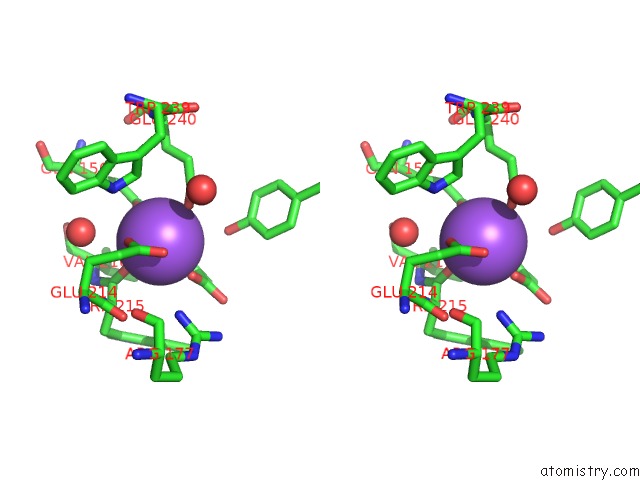
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structures of A Sodium-Alpha-Keto Acid Binding Subunit From A Trap Transporter in Its Closed Forms within 5.0Å range:
|
Reference:
S.Gonin,
P.Arnoux,
B.Pierru,
J.Lavergne,
B.Alonso,
M.Sabaty,
D.Pignol.
Crystal Structures of An Extracytoplasmic Solute Receptor From A Trap Transporter in Its Open and Closed Forms Reveal A Helix-Swapped Dimer Requiring A Cation For Alpha-Keto Acid Binding. Bmc Struct.Biol. V. 7 11 2007.
ISSN: ESSN 1472-6807
PubMed: 17362499
DOI: 10.1186/1472-6807-7-11
Page generated: Mon Oct 7 02:49:12 2024
ISSN: ESSN 1472-6807
PubMed: 17362499
DOI: 10.1186/1472-6807-7-11
Last articles
Ca in 5LOWCa in 5LPX
Ca in 5LK5
Ca in 5LPA
Ca in 5LOB
Ca in 5LP6
Ca in 5LO8
Ca in 5LOV
Ca in 5LIF
Ca in 5LJ4