Sodium »
PDB 2gg3-2gu7 »
2gu5 »
Sodium in PDB 2gu5: E. Coli Methionine Aminopeptidase in Complex with Nlep, 1: 1, Di-Metalated
Enzymatic activity of E. Coli Methionine Aminopeptidase in Complex with Nlep, 1: 1, Di-Metalated
All present enzymatic activity of E. Coli Methionine Aminopeptidase in Complex with Nlep, 1: 1, Di-Metalated:
3.4.11.18;
3.4.11.18;
Protein crystallography data
The structure of E. Coli Methionine Aminopeptidase in Complex with Nlep, 1: 1, Di-Metalated, PDB code: 2gu5
was solved by
Q.Z.Ye,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 14.19 / 1.60 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 49.790, 64.606, 76.354, 90.00, 107.77, 90.00 |
| R / Rfree (%) | 21.1 / 23.3 |
Other elements in 2gu5:
The structure of E. Coli Methionine Aminopeptidase in Complex with Nlep, 1: 1, Di-Metalated also contains other interesting chemical elements:
| Manganese | (Mn) | 4 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the E. Coli Methionine Aminopeptidase in Complex with Nlep, 1: 1, Di-Metalated
(pdb code 2gu5). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the E. Coli Methionine Aminopeptidase in Complex with Nlep, 1: 1, Di-Metalated, PDB code: 2gu5:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the E. Coli Methionine Aminopeptidase in Complex with Nlep, 1: 1, Di-Metalated, PDB code: 2gu5:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 2gu5
Go back to
Sodium binding site 1 out
of 2 in the E. Coli Methionine Aminopeptidase in Complex with Nlep, 1: 1, Di-Metalated
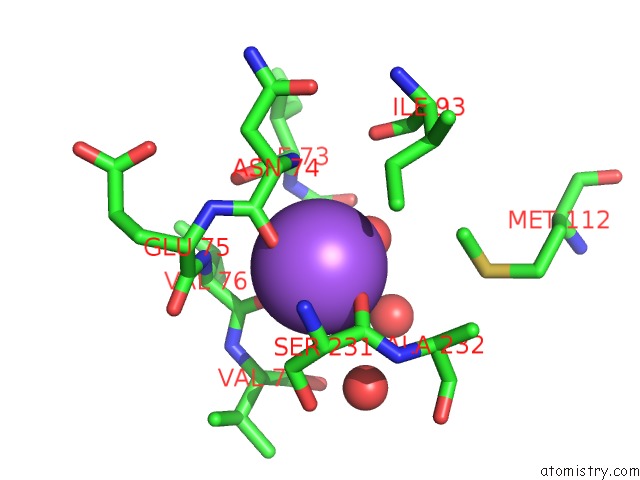
Mono view
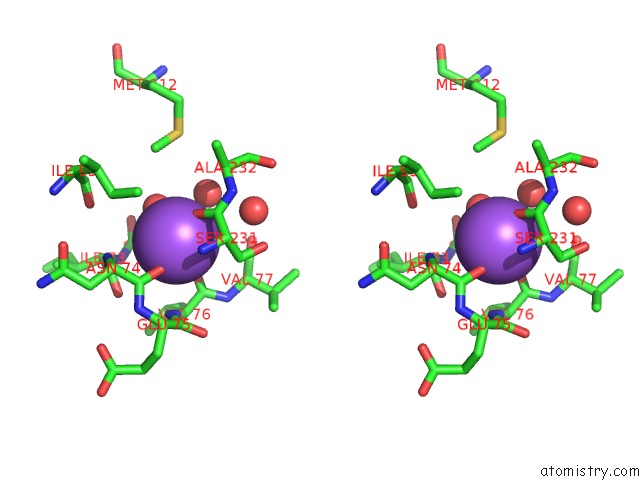
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of E. Coli Methionine Aminopeptidase in Complex with Nlep, 1: 1, Di-Metalated within 5.0Å range:
|
Sodium binding site 2 out of 2 in 2gu5
Go back to
Sodium binding site 2 out
of 2 in the E. Coli Methionine Aminopeptidase in Complex with Nlep, 1: 1, Di-Metalated
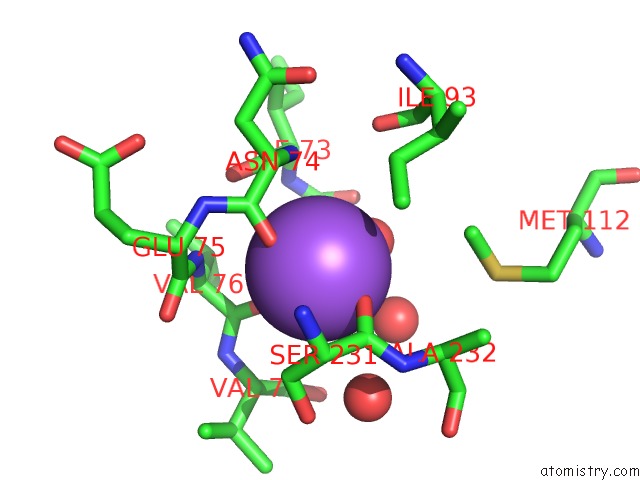
Mono view
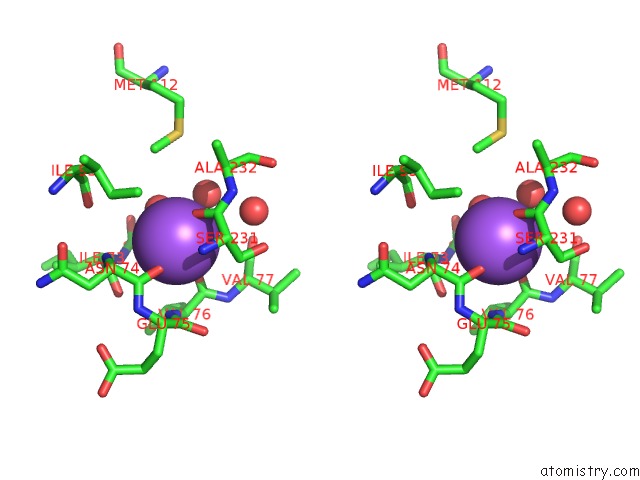
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of E. Coli Methionine Aminopeptidase in Complex with Nlep, 1: 1, Di-Metalated within 5.0Å range:
|
Reference:
Q.Z.Ye,
S.X.Xie,
Z.Q.Ma,
M.Huang,
R.P.Hanzlik.
Structural Basis of Catalysis By Monometalated Methionine Aminopeptidase. Proc.Natl.Acad.Sci.Usa V. 103 9470 2006.
ISSN: ISSN 0027-8424
PubMed: 16769889
DOI: 10.1073/PNAS.0602433103
Page generated: Mon Oct 7 02:41:43 2024
ISSN: ISSN 0027-8424
PubMed: 16769889
DOI: 10.1073/PNAS.0602433103
Last articles
F in 4DXHF in 4DXD
F in 4DVX
F in 4DVW
F in 4DTC
F in 4DVV
F in 4DVT
F in 4DV8
F in 4DDY
F in 4DUS