Sodium »
PDB 2czs-2e4r »
2dvo »
Sodium in PDB 2dvo: Structure of PH1917 Protein with the Complex of Itp From Pyrococcus Horikoshii
Protein crystallography data
The structure of Structure of PH1917 Protein with the Complex of Itp From Pyrococcus Horikoshii, PDB code: 2dvo
was solved by
N.K.Lokanath,
N.Kunishima,
Riken Structuralgenomics/Proteomics Initiative (Rsgi),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 2.21 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 78.261, 93.068, 54.051, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.5 / 22.9 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Structure of PH1917 Protein with the Complex of Itp From Pyrococcus Horikoshii
(pdb code 2dvo). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Structure of PH1917 Protein with the Complex of Itp From Pyrococcus Horikoshii, PDB code: 2dvo:
In total only one binding site of Sodium was determined in the Structure of PH1917 Protein with the Complex of Itp From Pyrococcus Horikoshii, PDB code: 2dvo:
Sodium binding site 1 out of 1 in 2dvo
Go back to
Sodium binding site 1 out
of 1 in the Structure of PH1917 Protein with the Complex of Itp From Pyrococcus Horikoshii
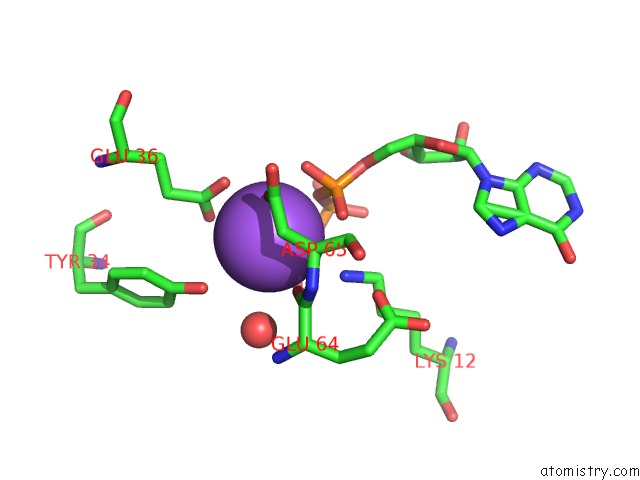
Mono view
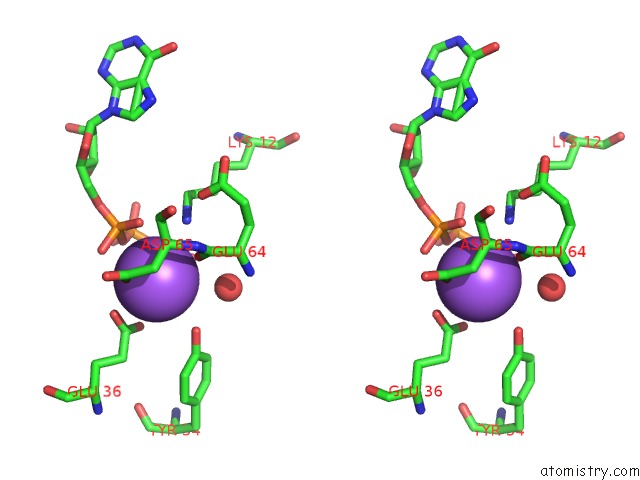
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Structure of PH1917 Protein with the Complex of Itp From Pyrococcus Horikoshii within 5.0Å range:
|
Reference:
N.K.Lokanath,
K.J.Pampa,
K.Takio,
N.Kunishima.
Structures of Dimeric Nonstandard Nucleotide Triphosphate Pyrophosphatase From Pyrococcus Horikoshii OT3: Functional Significance of Interprotomer Conformational Changes J.Mol.Biol. V. 375 1013 2008.
ISSN: ISSN 0022-2836
PubMed: 18062990
DOI: 10.1016/J.JMB.2007.11.018
Page generated: Mon Oct 7 02:16:08 2024
ISSN: ISSN 0022-2836
PubMed: 18062990
DOI: 10.1016/J.JMB.2007.11.018
Last articles
Cl in 5EOECl in 5EOU
Cl in 5EOO
Cl in 5EOI
Cl in 5ENJ
Cl in 5EN3
Cl in 5ENC
Cl in 5ENI
Cl in 5EMY
Cl in 5EN2