Sodium »
PDB 2aoe-2bhp »
2bhc »
Sodium in PDB 2bhc: Na Substituted E. Coli Aminopeptidase P
Enzymatic activity of Na Substituted E. Coli Aminopeptidase P
All present enzymatic activity of Na Substituted E. Coli Aminopeptidase P:
3.4.11.9;
3.4.11.9;
Protein crystallography data
The structure of Na Substituted E. Coli Aminopeptidase P, PDB code: 2bhc
was solved by
S.C.Graham,
C.S.Bond,
H.C.Freeman,
J.M.Guss,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 60.19 / 2.40 |
| Space group | I 41 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 138.783, 138.783, 231.012, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.6 / 20.2 |
Other elements in 2bhc:
The structure of Na Substituted E. Coli Aminopeptidase P also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Sodium Binding Sites:
The binding sites of Sodium atom in the Na Substituted E. Coli Aminopeptidase P
(pdb code 2bhc). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Na Substituted E. Coli Aminopeptidase P, PDB code: 2bhc:
In total only one binding site of Sodium was determined in the Na Substituted E. Coli Aminopeptidase P, PDB code: 2bhc:
Sodium binding site 1 out of 1 in 2bhc
Go back to
Sodium binding site 1 out
of 1 in the Na Substituted E. Coli Aminopeptidase P
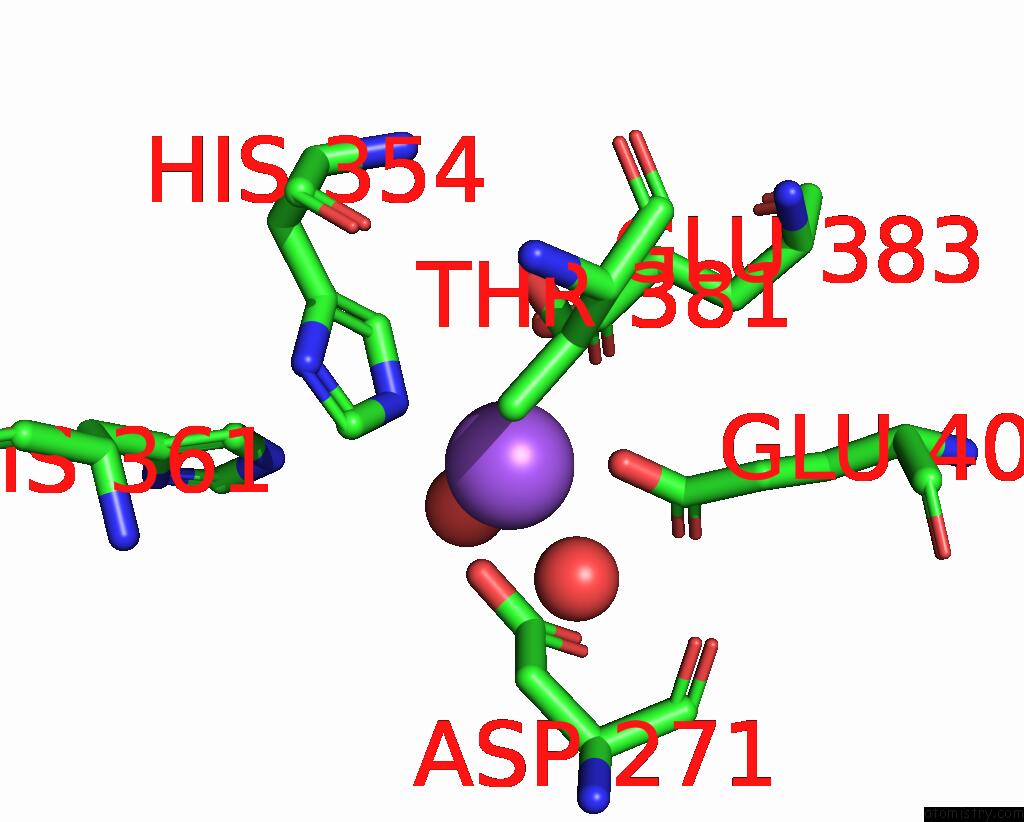
Mono view
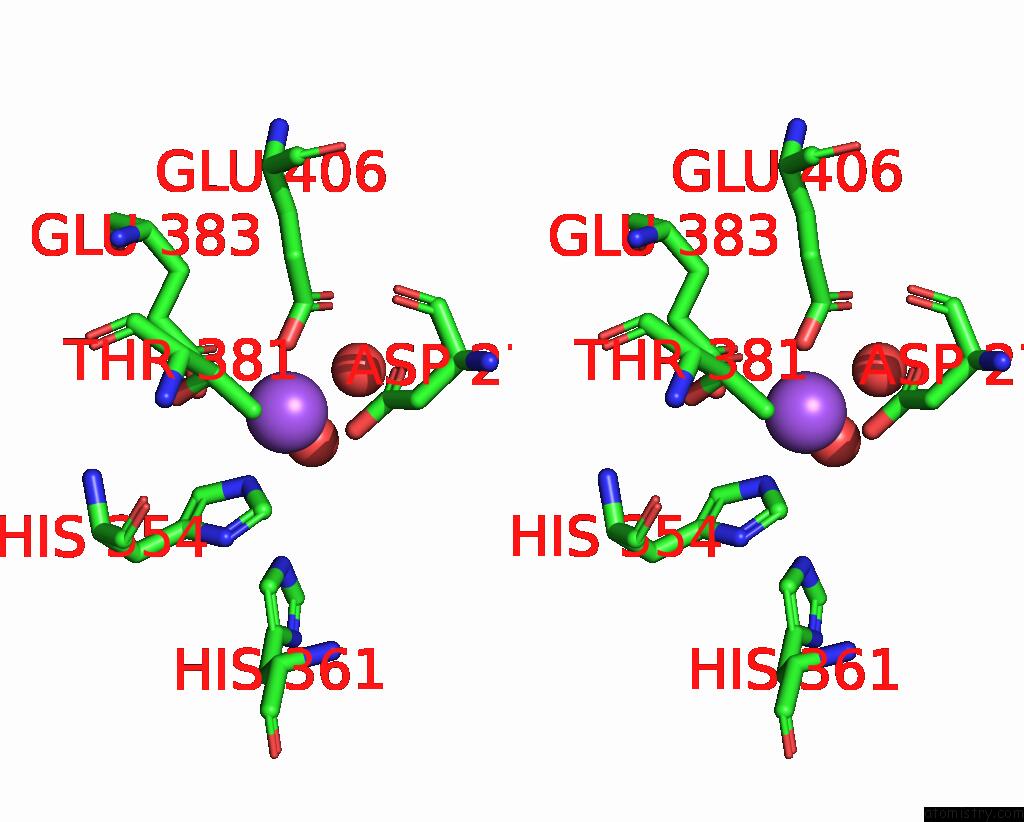
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Na Substituted E. Coli Aminopeptidase P within 5.0Å range:
|
Reference:
S.C.Graham,
C.S.Bond,
H.C.Freeman,
J.M.Guss.
Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, A Metalloprotease with A Dinuclear Metal Center. Biochemistry V. 44 13820 2005.
ISSN: ISSN 0006-2960
PubMed: 16229471
DOI: 10.1021/BI0512849
Page generated: Mon Oct 7 01:59:13 2024
ISSN: ISSN 0006-2960
PubMed: 16229471
DOI: 10.1021/BI0512849
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO