Sodium »
PDB 1s45-1t3p »
1sue »
Sodium in PDB 1sue: Subtilisin Bpn' From Bacillus Amyloliquefaciens, Mutant
Enzymatic activity of Subtilisin Bpn' From Bacillus Amyloliquefaciens, Mutant
All present enzymatic activity of Subtilisin Bpn' From Bacillus Amyloliquefaciens, Mutant:
3.4.21.62;
3.4.21.62;
Protein crystallography data
The structure of Subtilisin Bpn' From Bacillus Amyloliquefaciens, Mutant, PDB code: 1sue
was solved by
D.T.Gallagher,
P.Bryan,
Q.Pan,
G.L.Gilliland,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 16.00 / 1.80 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 45.590, 58.090, 86.580, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17 / n/a |
Sodium Binding Sites:
The binding sites of Sodium atom in the Subtilisin Bpn' From Bacillus Amyloliquefaciens, Mutant
(pdb code 1sue). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Subtilisin Bpn' From Bacillus Amyloliquefaciens, Mutant, PDB code: 1sue:
In total only one binding site of Sodium was determined in the Subtilisin Bpn' From Bacillus Amyloliquefaciens, Mutant, PDB code: 1sue:
Sodium binding site 1 out of 1 in 1sue
Go back to
Sodium binding site 1 out
of 1 in the Subtilisin Bpn' From Bacillus Amyloliquefaciens, Mutant
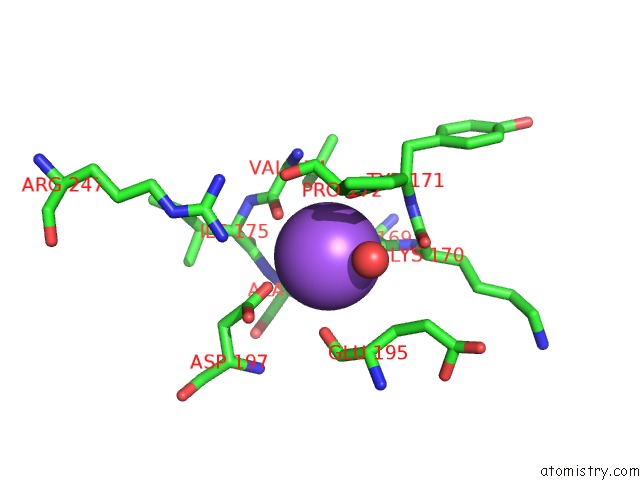
Mono view
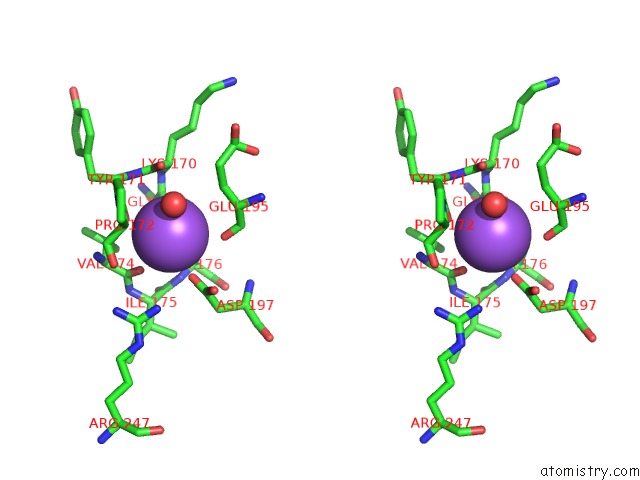
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Subtilisin Bpn' From Bacillus Amyloliquefaciens, Mutant within 5.0Å range:
|
Reference:
D.T.Gallagher,
Q.W.Pan,
G.L.Gilliland.
Mechanism of Ionic Strength Dependence of Crystal Growth Rates in A Subtilisin Variant. J.Cryst.Growth V. 193 665 1998.
ISSN: ISSN 0022-0248
Page generated: Sun Aug 17 07:44:53 2025
ISSN: ISSN 0022-0248
Last articles
Na in 3L8LNa in 3L8W
Na in 3L8M
Na in 3L25
Na in 3L8G
Na in 3L28
Na in 3L7X
Na in 3L6T
Na in 3L6I
Na in 3L27