Sodium »
PDB 1s45-1t3p »
1s5b »
Sodium in PDB 1s5b: Cholera Holotoxin with An A-Subunit Y30S Mutation Form 3
Enzymatic activity of Cholera Holotoxin with An A-Subunit Y30S Mutation Form 3
All present enzymatic activity of Cholera Holotoxin with An A-Subunit Y30S Mutation Form 3:
2.4.2.36;
2.4.2.36;
Protein crystallography data
The structure of Cholera Holotoxin with An A-Subunit Y30S Mutation Form 3, PDB code: 1s5b
was solved by
C.J.O'neal,
E.I.Amaya,
M.G.Jobling,
R.K.Holmes,
W.G.Hol,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.02 / 2.13 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 60.104, 107.431, 65.950, 90.00, 91.29, 90.00 |
| R / Rfree (%) | 17.7 / 22.4 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Cholera Holotoxin with An A-Subunit Y30S Mutation Form 3
(pdb code 1s5b). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Cholera Holotoxin with An A-Subunit Y30S Mutation Form 3, PDB code: 1s5b:
In total only one binding site of Sodium was determined in the Cholera Holotoxin with An A-Subunit Y30S Mutation Form 3, PDB code: 1s5b:
Sodium binding site 1 out of 1 in 1s5b
Go back to
Sodium binding site 1 out
of 1 in the Cholera Holotoxin with An A-Subunit Y30S Mutation Form 3
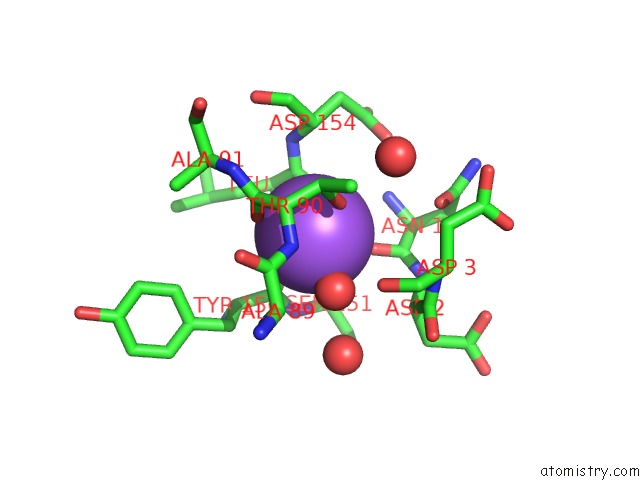
Mono view
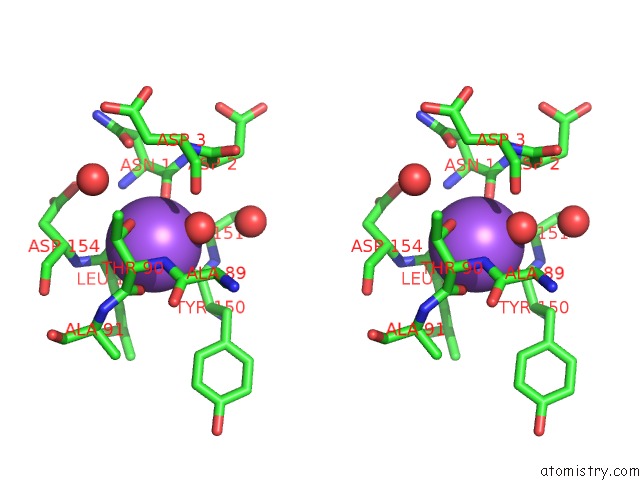
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Cholera Holotoxin with An A-Subunit Y30S Mutation Form 3 within 5.0Å range:
|
Reference:
C.J.O'neal,
E.I.Amaya,
M.G.Jobling,
R.K.Holmes,
W.G.Hol.
Crystal Structures of An Intrinsically Active Cholera Toxin Mutant Yield Insight Into the Toxin Activation Mechanism Biochemistry V. 43 3772 2004.
ISSN: ISSN 0006-2960
PubMed: 15049684
DOI: 10.1021/BI0360152
Page generated: Sun Aug 17 07:41:14 2025
ISSN: ISSN 0006-2960
PubMed: 15049684
DOI: 10.1021/BI0360152
Last articles
Na in 5MURNa in 5MXB
Na in 5MTP
Na in 5MSS
Na in 5MU9
Na in 5MTO
Na in 5MSE
Na in 5MOZ
Na in 5MRT
Na in 5MRR