Sodium »
PDB 1ph7-1qsx »
1qch »
Sodium in PDB 1qch: Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution
Sodium Binding Sites:
The binding sites of Sodium atom in the Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution
(pdb code 1qch). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 9 binding sites of Sodium where determined in the Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution, PDB code: 1qch:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
In total 9 binding sites of Sodium where determined in the Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution, PDB code: 1qch:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
Sodium binding site 1 out of 9 in 1qch
Go back to
Sodium binding site 1 out
of 9 in the Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution within 5.0Å range:
|
Sodium binding site 2 out of 9 in 1qch
Go back to
Sodium binding site 2 out
of 9 in the Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution within 5.0Å range:
|
Sodium binding site 3 out of 9 in 1qch
Go back to
Sodium binding site 3 out
of 9 in the Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution

Mono view
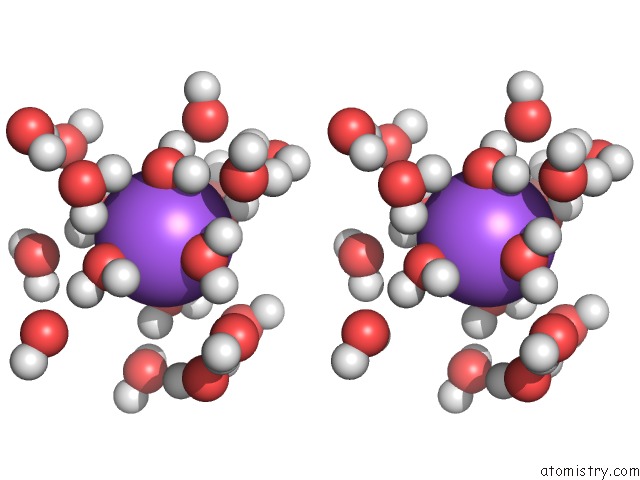
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution within 5.0Å range:
|
Sodium binding site 4 out of 9 in 1qch
Go back to
Sodium binding site 4 out
of 9 in the Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution within 5.0Å range:
|
Sodium binding site 5 out of 9 in 1qch
Go back to
Sodium binding site 5 out
of 9 in the Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 5 of Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution within 5.0Å range:
|
Sodium binding site 6 out of 9 in 1qch
Go back to
Sodium binding site 6 out
of 9 in the Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 6 of Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution within 5.0Å range:
|
Sodium binding site 7 out of 9 in 1qch
Go back to
Sodium binding site 7 out
of 9 in the Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 7 of Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution within 5.0Å range:
|
Sodium binding site 8 out of 9 in 1qch
Go back to
Sodium binding site 8 out
of 9 in the Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 8 of Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution within 5.0Å range:
|
Sodium binding site 9 out of 9 in 1qch
Go back to
Sodium binding site 9 out
of 9 in the Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution

Mono view
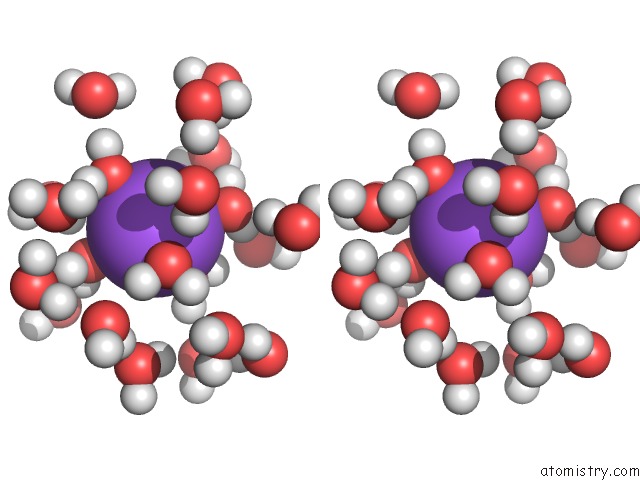
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 9 of Structure, Dynamics and Hydration of the Nogalamycin- D(Atgcat)2 Complex Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution within 5.0Å range:
|
Reference:
H.E.Williams,
M.S.Searle.
Structure, Dynamics and Hydration of the Nogalamycin-D(Atgcat)2COMPLEX Determined By uc(Nmr) and Molecular Dynamics Simulations in Solution. J.Mol.Biol. V. 290 699 1999.
ISSN: ISSN 0022-2836
PubMed: 10395824
DOI: 10.1006/JMBI.1999.2903
Page generated: Sun Oct 6 21:25:30 2024
ISSN: ISSN 0022-2836
PubMed: 10395824
DOI: 10.1006/JMBI.1999.2903
Last articles
Ca in 5N0KCa in 5MYI
Ca in 5MZO
Ca in 5MXX
Ca in 5MY9
Ca in 5MYC
Ca in 5MWC
Ca in 5MWB
Ca in 5MWK
Ca in 5MW5