Sodium »
PDB 1m90-1nji »
1moj »
Sodium in PDB 1moj: Crystal Structure of An Archaeal Dps-Homologue From Halobacterium Salinarum
Protein crystallography data
The structure of Crystal Structure of An Archaeal Dps-Homologue From Halobacterium Salinarum, PDB code: 1moj
was solved by
K.Zeth,
S.Offermann,
L.O.Essen,
D.Oesterhelt,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.88 / 1.90 |
| Space group | P 3 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 90.747, 90.747, 149.410, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 14.7 / 18.2 |
Other elements in 1moj:
The structure of Crystal Structure of An Archaeal Dps-Homologue From Halobacterium Salinarum also contains other interesting chemical elements:
| Magnesium | (Mg) | 4 atoms |
| Iron | (Fe) | 6 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of An Archaeal Dps-Homologue From Halobacterium Salinarum
(pdb code 1moj). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Crystal Structure of An Archaeal Dps-Homologue From Halobacterium Salinarum, PDB code: 1moj:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Crystal Structure of An Archaeal Dps-Homologue From Halobacterium Salinarum, PDB code: 1moj:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 1moj
Go back to
Sodium binding site 1 out
of 2 in the Crystal Structure of An Archaeal Dps-Homologue From Halobacterium Salinarum
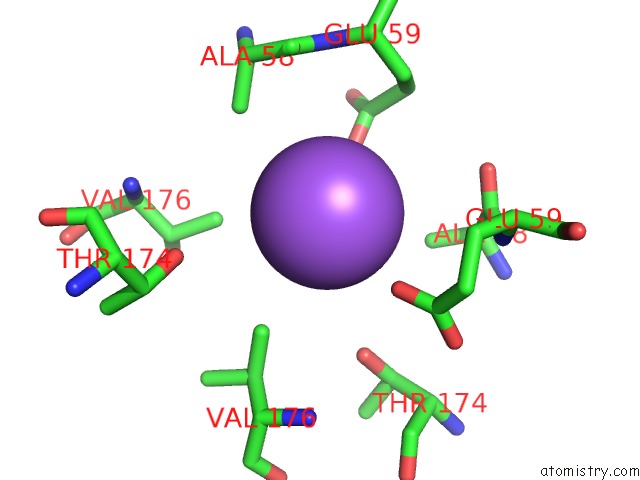
Mono view
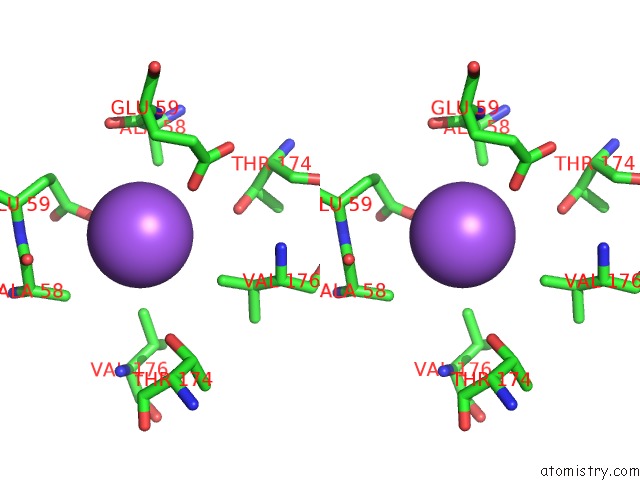
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of An Archaeal Dps-Homologue From Halobacterium Salinarum within 5.0Å range:
|
Sodium binding site 2 out of 2 in 1moj
Go back to
Sodium binding site 2 out
of 2 in the Crystal Structure of An Archaeal Dps-Homologue From Halobacterium Salinarum
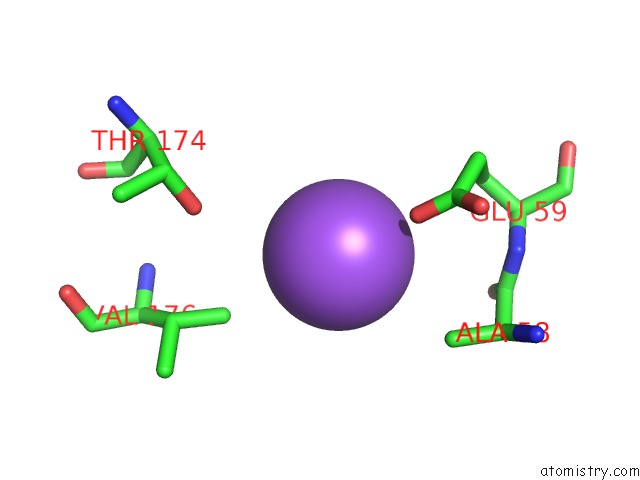
Mono view
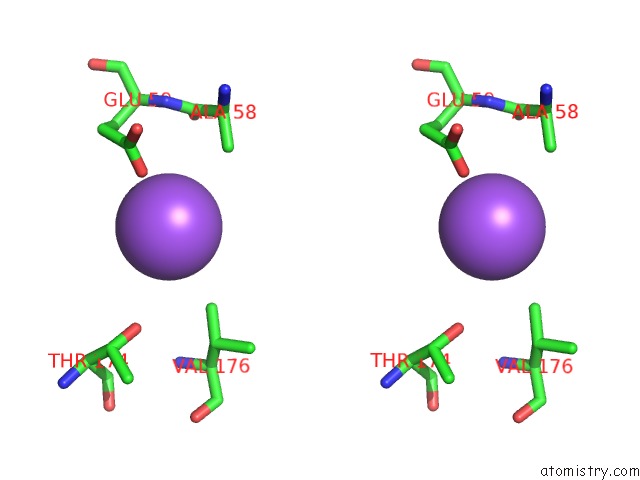
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure of An Archaeal Dps-Homologue From Halobacterium Salinarum within 5.0Å range:
|
Reference:
K.Zeth,
S.Offermann,
L.O.Essen,
D.Oesterhelt.
Iron-Oxo Clusters Biomineralizing on Protein Surfaces: Structural Analysis of Halobacterium Salinarum Dpsa in Its Low- and High-Iron States. Proc.Natl.Acad.Sci.Usa V. 101 13780 2004.
ISSN: ISSN 0027-8424
PubMed: 15365182
DOI: 10.1073/PNAS.0401821101
Page generated: Sun Oct 6 20:33:53 2024
ISSN: ISSN 0027-8424
PubMed: 15365182
DOI: 10.1073/PNAS.0401821101
Last articles
Cl in 5RAJCl in 5RAM
Cl in 5RAL
Cl in 5RAK
Cl in 5RAF
Cl in 5RAG
Cl in 5RAH
Cl in 5RAI
Cl in 5RAE
Cl in 5RAD