Sodium »
PDB 1k7e-1l0i »
1kfe »
Sodium in PDB 1kfe: Crystal Structure of ALPHAT183V Mutant of Tryptophan Synthase From Salmonella Typhimurium with L-Ser Bound to the Beta Site
Protein crystallography data
The structure of Crystal Structure of ALPHAT183V Mutant of Tryptophan Synthase From Salmonella Typhimurium with L-Ser Bound to the Beta Site, PDB code: 1kfe
was solved by
V.Kulik,
M.Weyand,
R.Siedel,
D.Niks,
D.Arac,
M.F.Dunn,
I.Schlichting,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.97 / 1.75 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 182.224, 59.985, 67.072, 90.00, 94.68, 90.00 |
| R / Rfree (%) | 19.2 / 22.5 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of ALPHAT183V Mutant of Tryptophan Synthase From Salmonella Typhimurium with L-Ser Bound to the Beta Site
(pdb code 1kfe). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Crystal Structure of ALPHAT183V Mutant of Tryptophan Synthase From Salmonella Typhimurium with L-Ser Bound to the Beta Site, PDB code: 1kfe:
In total only one binding site of Sodium was determined in the Crystal Structure of ALPHAT183V Mutant of Tryptophan Synthase From Salmonella Typhimurium with L-Ser Bound to the Beta Site, PDB code: 1kfe:
Sodium binding site 1 out of 1 in 1kfe
Go back to
Sodium binding site 1 out
of 1 in the Crystal Structure of ALPHAT183V Mutant of Tryptophan Synthase From Salmonella Typhimurium with L-Ser Bound to the Beta Site
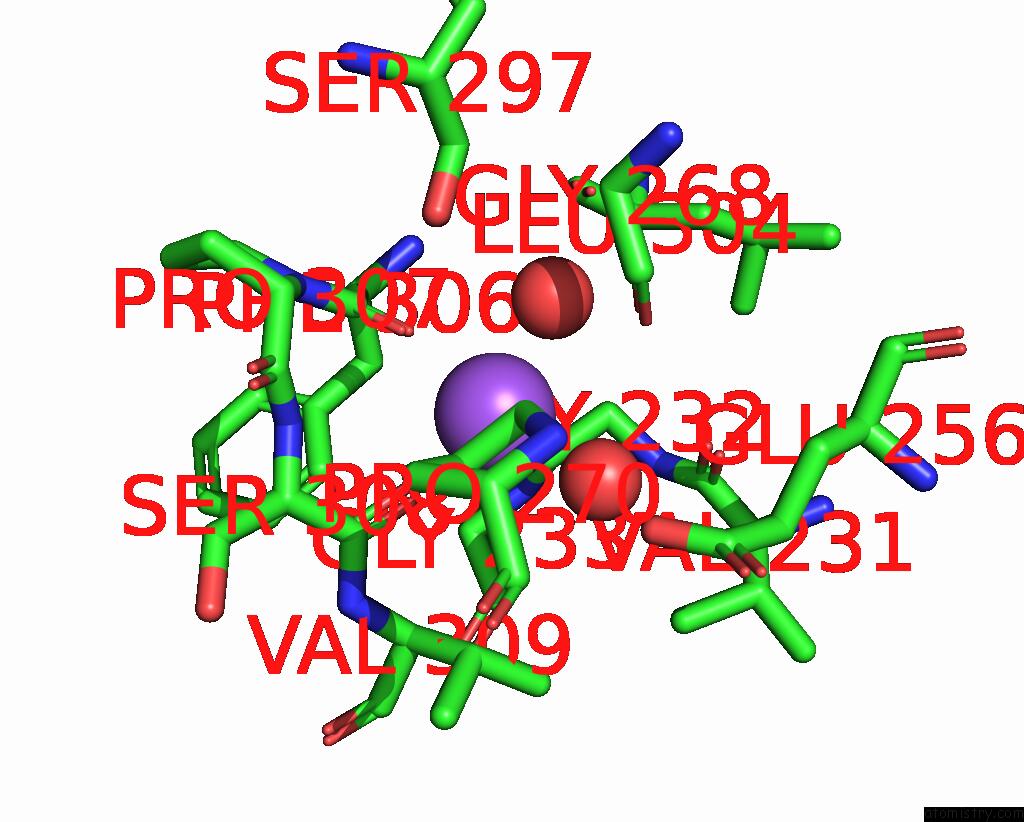
Mono view
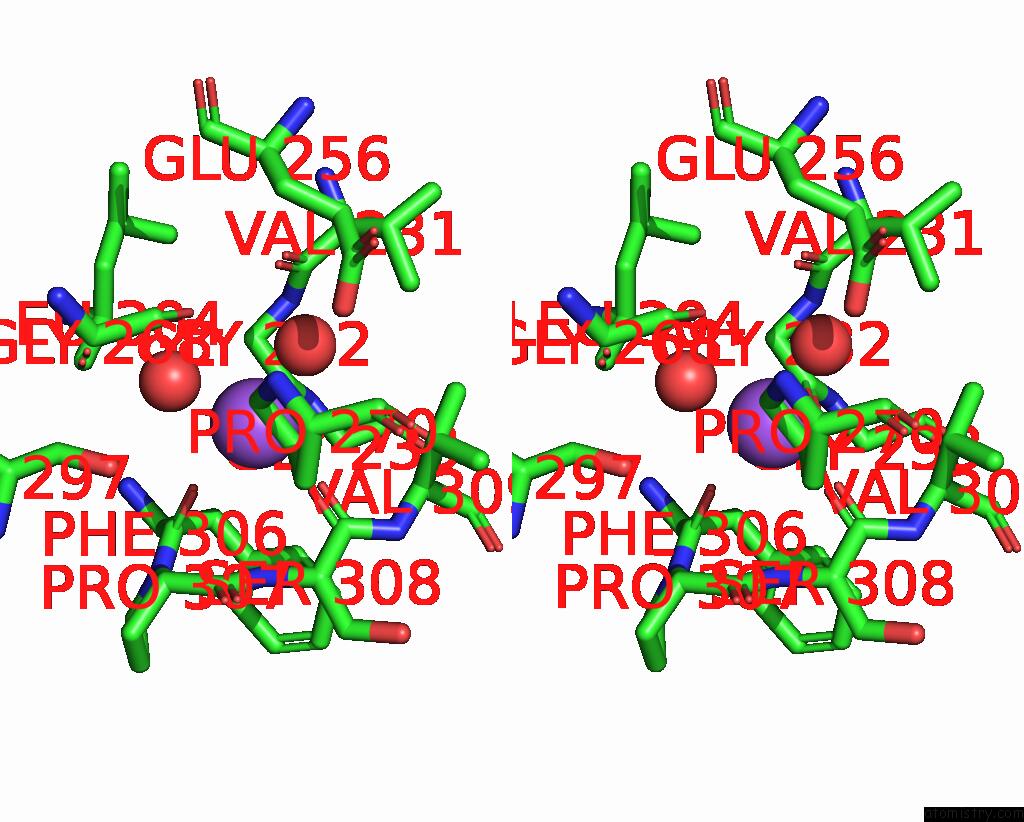
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of ALPHAT183V Mutant of Tryptophan Synthase From Salmonella Typhimurium with L-Ser Bound to the Beta Site within 5.0Å range:
|
Reference:
V.Kulik,
M.Weyand,
R.Siedel,
D.Niks,
D.Arac,
M.F.Dunn,
I.Schlichting.
On the Role of ALPHATHR183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase J.Mol.Biol. V. 324 677 2002.
ISSN: ISSN 0022-2836
PubMed: 12460570
DOI: 10.1016/S0022-2836(02)01109-9
Page generated: Sun Aug 17 06:06:31 2025
ISSN: ISSN 0022-2836
PubMed: 12460570
DOI: 10.1016/S0022-2836(02)01109-9
Last articles
Na in 3I04Na in 3I01
Na in 3I0X
Na in 3I0W
Na in 3HWX
Na in 3HZN
Na in 3HZO
Na in 3HYS
Na in 3HYJ
Na in 3HYA