Sodium »
PDB 1evr-1gb5 »
1fuy »
Sodium in PDB 1fuy: Crystal Structure of BETAA169L/BETAC170W Double Mutant of Tryptophan Synthase Complexed with 5-Fluoro-Indole-Propanol Phosphate
Enzymatic activity of Crystal Structure of BETAA169L/BETAC170W Double Mutant of Tryptophan Synthase Complexed with 5-Fluoro-Indole-Propanol Phosphate
All present enzymatic activity of Crystal Structure of BETAA169L/BETAC170W Double Mutant of Tryptophan Synthase Complexed with 5-Fluoro-Indole-Propanol Phosphate:
4.2.1.20;
4.2.1.20;
Protein crystallography data
The structure of Crystal Structure of BETAA169L/BETAC170W Double Mutant of Tryptophan Synthase Complexed with 5-Fluoro-Indole-Propanol Phosphate, PDB code: 1fuy
was solved by
M.Weyand,
I.Schlichting,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 2.25 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 184.000, 60.700, 67.400, 90.00, 95.75, 90.00 |
| R / Rfree (%) | 17 / 22.4 |
Other elements in 1fuy:
The structure of Crystal Structure of BETAA169L/BETAC170W Double Mutant of Tryptophan Synthase Complexed with 5-Fluoro-Indole-Propanol Phosphate also contains other interesting chemical elements:
| Fluorine | (F) | 1 atom |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of BETAA169L/BETAC170W Double Mutant of Tryptophan Synthase Complexed with 5-Fluoro-Indole-Propanol Phosphate
(pdb code 1fuy). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Crystal Structure of BETAA169L/BETAC170W Double Mutant of Tryptophan Synthase Complexed with 5-Fluoro-Indole-Propanol Phosphate, PDB code: 1fuy:
In total only one binding site of Sodium was determined in the Crystal Structure of BETAA169L/BETAC170W Double Mutant of Tryptophan Synthase Complexed with 5-Fluoro-Indole-Propanol Phosphate, PDB code: 1fuy:
Sodium binding site 1 out of 1 in 1fuy
Go back to
Sodium binding site 1 out
of 1 in the Crystal Structure of BETAA169L/BETAC170W Double Mutant of Tryptophan Synthase Complexed with 5-Fluoro-Indole-Propanol Phosphate
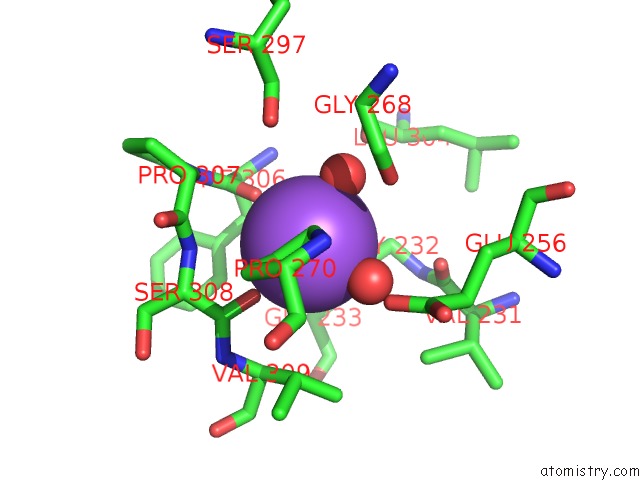
Mono view
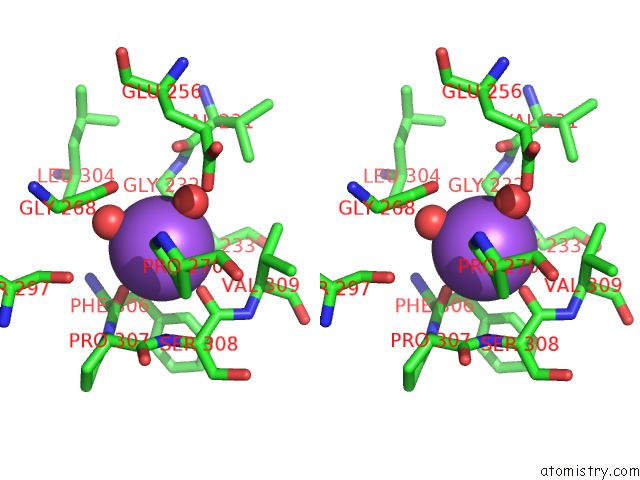
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of BETAA169L/BETAC170W Double Mutant of Tryptophan Synthase Complexed with 5-Fluoro-Indole-Propanol Phosphate within 5.0Å range:
|
Reference:
M.Weyand,
I.Schlichting.
Structural Basis For the Impaired Channeling and Allosteric Inter-Subunit Communication in the Beta A169L/Beta C170W Mutant of Tryptophan Synthase. J.Biol.Chem. V. 275 41058 2000.
ISSN: ISSN 0021-9258
PubMed: 11034989
DOI: 10.1074/JBC.C000479200
Page generated: Sun Oct 6 18:29:08 2024
ISSN: ISSN 0021-9258
PubMed: 11034989
DOI: 10.1074/JBC.C000479200
Last articles
Cl in 5QCCCl in 5QCF
Cl in 5QCD
Cl in 5QCA
Cl in 5QBY
Cl in 5QC8
Cl in 5QBV
Cl in 5QBZ
Cl in 5QBW
Cl in 5QB4