Sodium in PDB 8thj: Cryo-Em Structure of the Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer)
Sodium Binding Sites:
The binding sites of Sodium atom in the Cryo-Em Structure of the Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer)
(pdb code 8thj). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 4 binding sites of Sodium where determined in the Cryo-Em Structure of the Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer), PDB code: 8thj:
Jump to Sodium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Sodium where determined in the Cryo-Em Structure of the Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer), PDB code: 8thj:
Jump to Sodium binding site number: 1; 2; 3; 4;
Sodium binding site 1 out of 4 in 8thj
Go back to
Sodium binding site 1 out
of 4 in the Cryo-Em Structure of the Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer)

Mono view
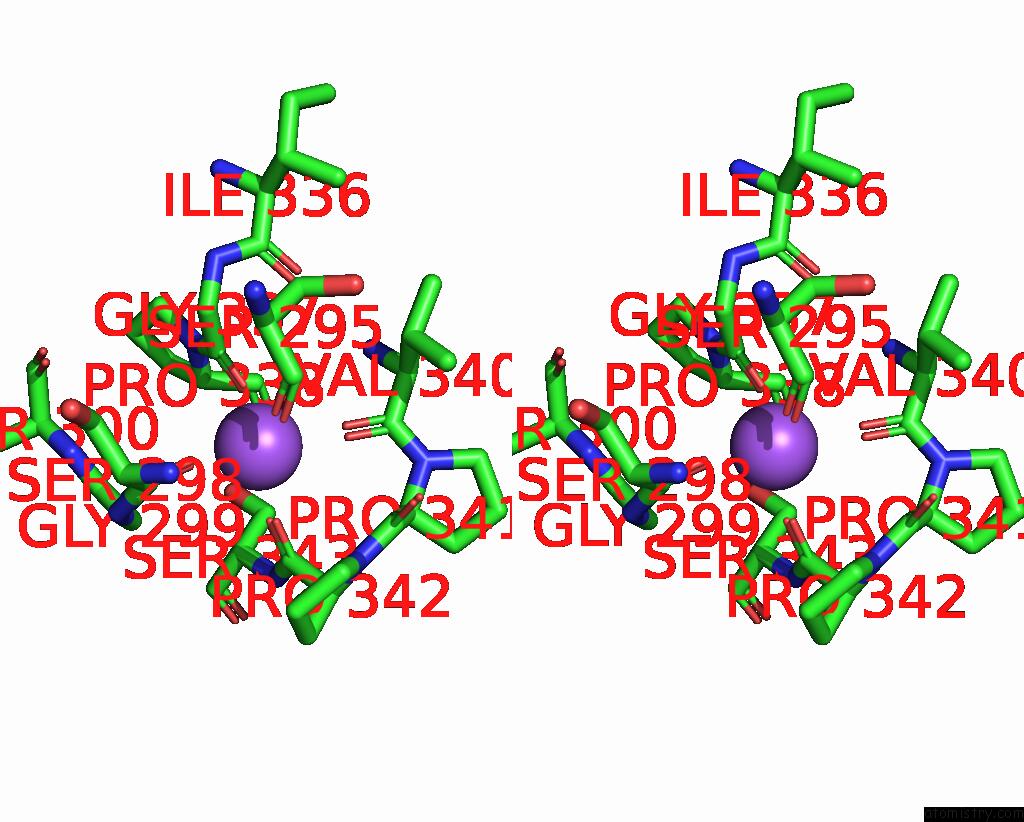
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Cryo-Em Structure of the Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer) within 5.0Å range:
|
Sodium binding site 2 out of 4 in 8thj
Go back to
Sodium binding site 2 out
of 4 in the Cryo-Em Structure of the Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer)
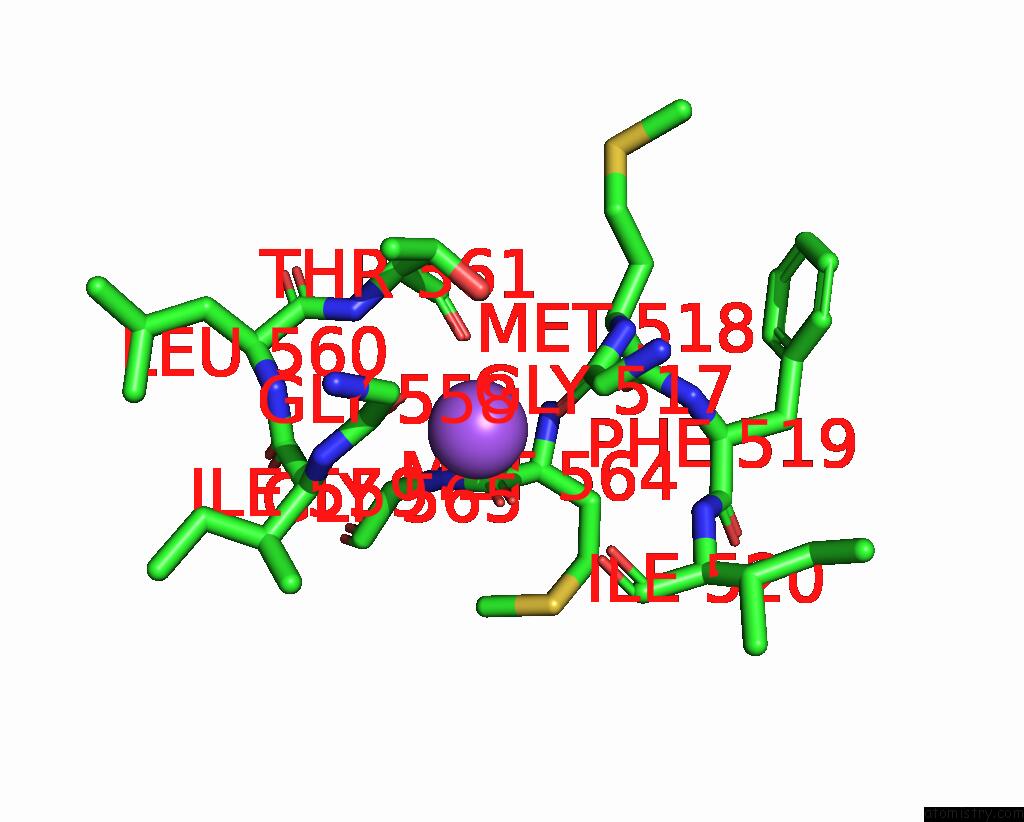
Mono view
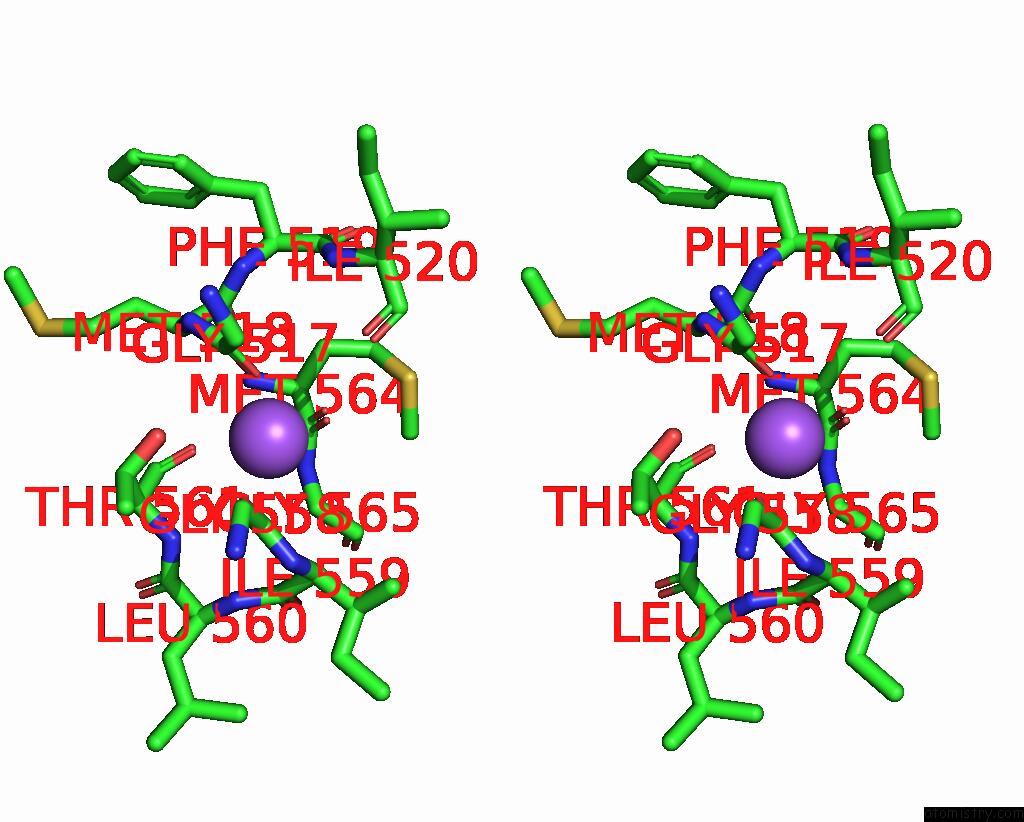
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Cryo-Em Structure of the Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer) within 5.0Å range:
|
Sodium binding site 3 out of 4 in 8thj
Go back to
Sodium binding site 3 out
of 4 in the Cryo-Em Structure of the Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer)
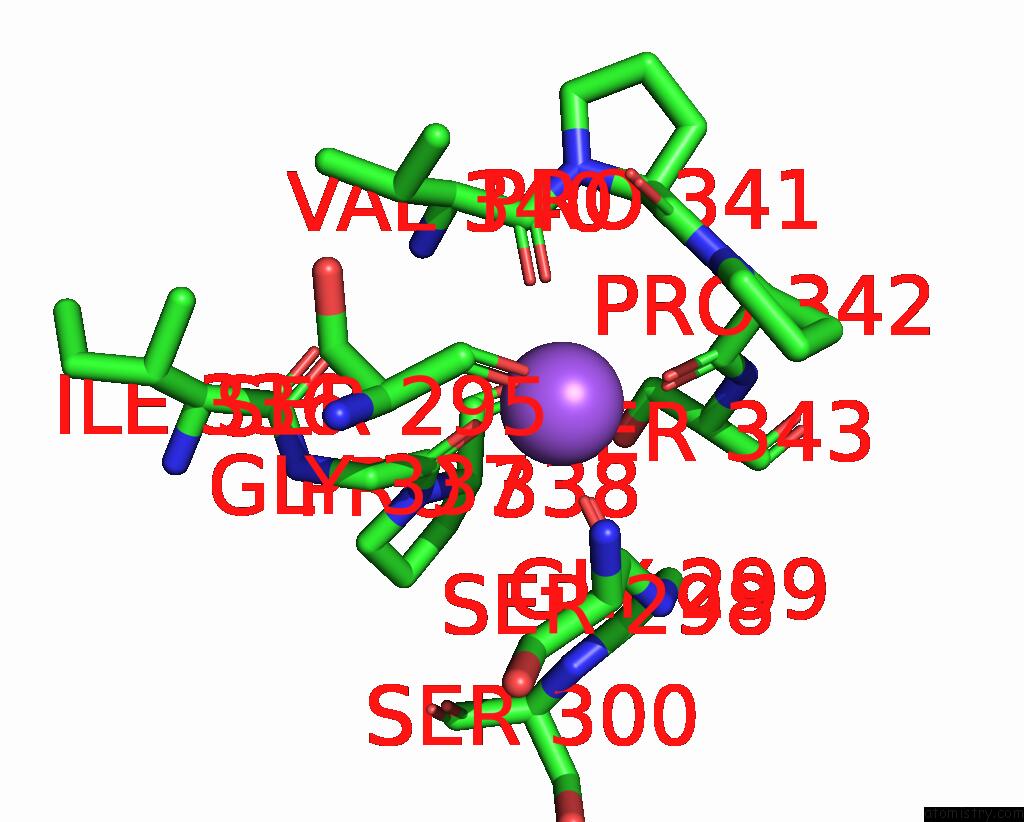
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Cryo-Em Structure of the Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer) within 5.0Å range:
|
Sodium binding site 4 out of 4 in 8thj
Go back to
Sodium binding site 4 out
of 4 in the Cryo-Em Structure of the Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer)
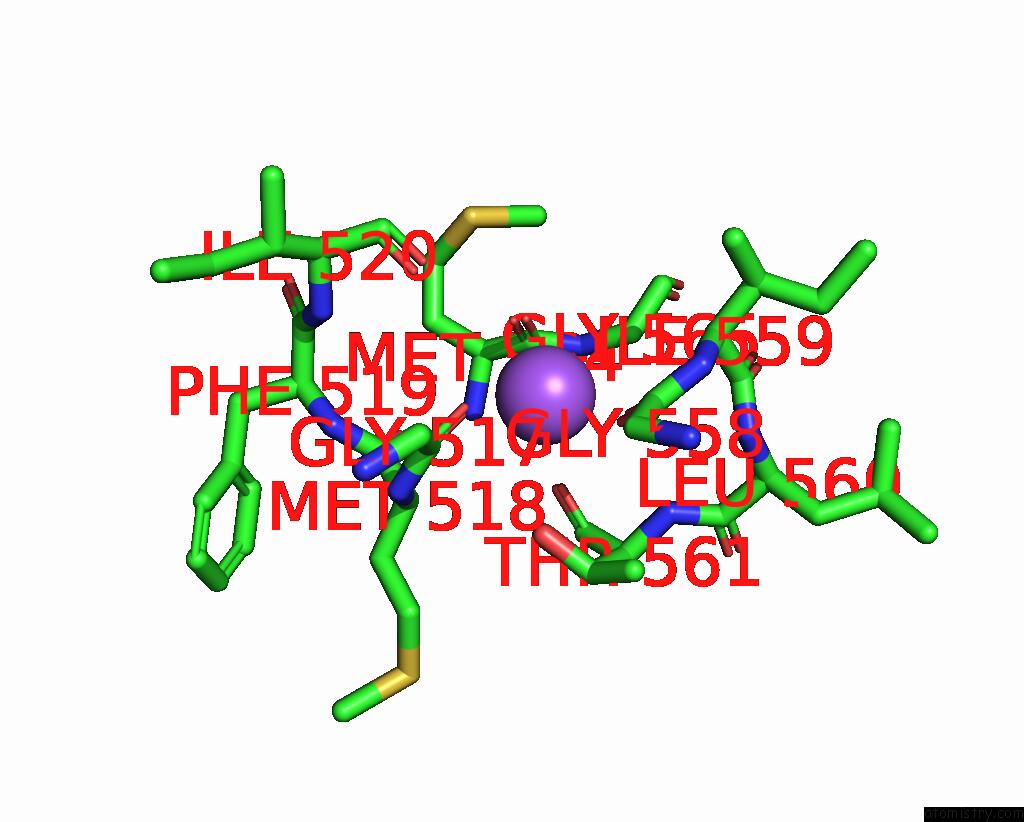
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Cryo-Em Structure of the Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer) within 5.0Å range:
|
Reference:
M.J.Currie,
J.S.Davies,
M.Scalise,
A.Gulati,
J.D.Wright,
M.C.Newton-Vesty,
G.S.Abeysekera,
R.Subramanian,
W.Y.Wahlgren,
R.Friemann,
J.R.Allison,
P.D.Mace,
M.D.W.Griffin,
B.Demeler,
S.Wakatsuki,
D.Drew,
C.Indiveri,
R.C.J.Dobson,
R.A.North.
Structural and Biophysical Analysis of A Haemophilus Influenzae Tripartite Atp-Independent Periplasmic (Trap) Transporter Elife 2023.
ISSN: ESSN 2050-084X
DOI: 10.7554/ELIFE.92307.1
Page generated: Wed Oct 9 13:40:18 2024
ISSN: ESSN 2050-084X
DOI: 10.7554/ELIFE.92307.1
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW