Sodium in PDB 7sq6: Cryo-Em Structure of Mouse Agonist Ml-SA1-Bound TRPML1 Channel at 2.32 Angstrom Resolution
Sodium Binding Sites:
The binding sites of Sodium atom in the Cryo-Em Structure of Mouse Agonist Ml-SA1-Bound TRPML1 Channel at 2.32 Angstrom Resolution
(pdb code 7sq6). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Cryo-Em Structure of Mouse Agonist Ml-SA1-Bound TRPML1 Channel at 2.32 Angstrom Resolution, PDB code: 7sq6:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Cryo-Em Structure of Mouse Agonist Ml-SA1-Bound TRPML1 Channel at 2.32 Angstrom Resolution, PDB code: 7sq6:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 7sq6
Go back to
Sodium binding site 1 out
of 2 in the Cryo-Em Structure of Mouse Agonist Ml-SA1-Bound TRPML1 Channel at 2.32 Angstrom Resolution
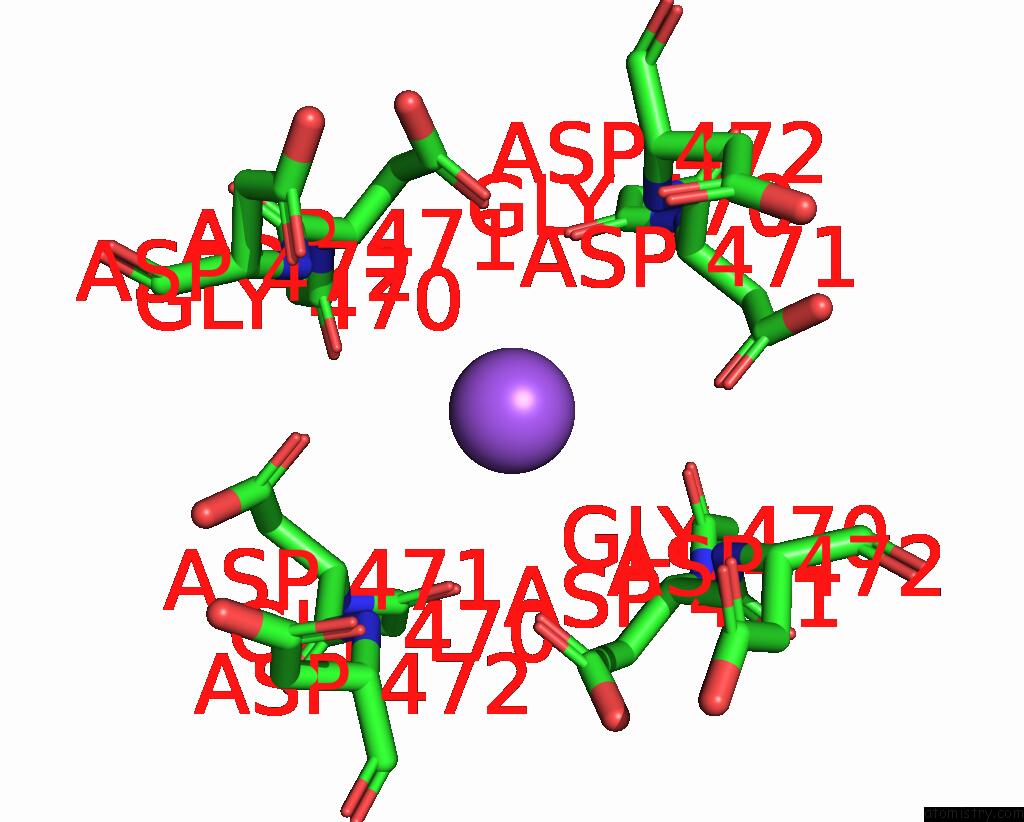
Mono view
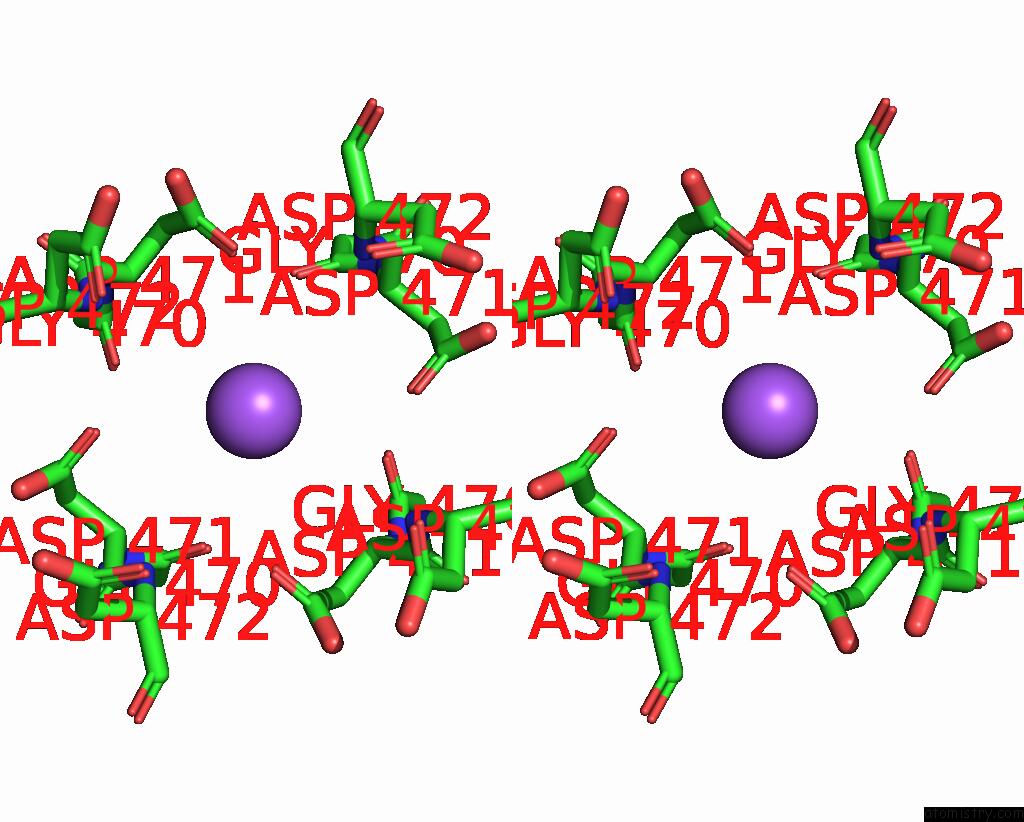
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Cryo-Em Structure of Mouse Agonist Ml-SA1-Bound TRPML1 Channel at 2.32 Angstrom Resolution within 5.0Å range:
|
Sodium binding site 2 out of 2 in 7sq6
Go back to
Sodium binding site 2 out
of 2 in the Cryo-Em Structure of Mouse Agonist Ml-SA1-Bound TRPML1 Channel at 2.32 Angstrom Resolution

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Cryo-Em Structure of Mouse Agonist Ml-SA1-Bound TRPML1 Channel at 2.32 Angstrom Resolution within 5.0Å range:
|
Reference:
N.Gan,
Y.Han,
W.Zeng,
Y.Wang,
J.Xue,
Y.Jiang.
Structural Mechanism of Allosteric Activation of TRPML1 By Pi(3,5)P 2 and Rapamycin. Proc.Natl.Acad.Sci.Usa V. 119 2022.
ISSN: ESSN 1091-6490
PubMed: 35131932
DOI: 10.1073/PNAS.2120404119
Page generated: Wed Oct 9 09:01:25 2024
ISSN: ESSN 1091-6490
PubMed: 35131932
DOI: 10.1073/PNAS.2120404119
Last articles
Zn in 9MJ5Zn in 9HNW
Zn in 9G0L
Zn in 9FNE
Zn in 9DZN
Zn in 9E0I
Zn in 9D32
Zn in 9DAK
Zn in 8ZXC
Zn in 8ZUF